
Pacific flying fox faeces associated circular DNA virus-13
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.12
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTV9|A0A140CTV9_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-13 OX=1796008 PE=4 SV=1
MM1 pKa = 7.33SVHH4 pKa = 7.06GNRR7 pKa = 11.84AWCFTLNNPTDD18 pKa = 3.84EE19 pKa = 4.69EE20 pKa = 4.41KK21 pKa = 11.15LLLPTSPEE29 pKa = 4.16SLPLWPRR36 pKa = 11.84CTALVYY42 pKa = 10.43QLEE45 pKa = 4.37EE46 pKa = 4.59GEE48 pKa = 4.55NGTPHH53 pKa = 5.5IQGYY57 pKa = 10.64ARR59 pKa = 11.84FDD61 pKa = 3.6RR62 pKa = 11.84QRR64 pKa = 11.84TLATVKK70 pKa = 10.02TILARR75 pKa = 11.84AHH77 pKa = 6.24WEE79 pKa = 3.7PAKK82 pKa = 10.92GSIQANLDD90 pKa = 3.43YY91 pKa = 10.87CGKK94 pKa = 9.58QLGRR98 pKa = 11.84LSEE101 pKa = 3.9PVIIGFGGLRR111 pKa = 11.84PGASGPSKK119 pKa = 9.99PHH121 pKa = 6.56LKK123 pKa = 10.02RR124 pKa = 11.84AQFVDD129 pKa = 6.11LIASSPDD136 pKa = 3.13VSIPEE141 pKa = 4.63LLDD144 pKa = 3.28QGALEE149 pKa = 4.36VLATQPNLLGTLRR162 pKa = 11.84GFLFQDD168 pKa = 3.76LRR170 pKa = 11.84RR171 pKa = 11.84DD172 pKa = 3.69GVIVDD177 pKa = 4.7LYY179 pKa = 11.35YY180 pKa = 11.39GLTGTGKK187 pKa = 10.18SRR189 pKa = 11.84LAFDD193 pKa = 4.69EE194 pKa = 4.39YY195 pKa = 10.65PGAFRR200 pKa = 11.84KK201 pKa = 8.41TSGSWWDD208 pKa = 3.48SYY210 pKa = 11.65AGEE213 pKa = 4.32LSVVLDD219 pKa = 4.46DD220 pKa = 5.71FDD222 pKa = 5.04DD223 pKa = 4.33AFMPIGDD230 pKa = 4.98LLRR233 pKa = 11.84TLDD236 pKa = 4.09RR237 pKa = 11.84YY238 pKa = 9.12PLRR241 pKa = 11.84MPVKK245 pKa = 10.28GGFIQLIANHH255 pKa = 6.76FIITSNHH262 pKa = 6.22LPSEE266 pKa = 4.88WYY268 pKa = 9.87PNAPPCRR275 pKa = 11.84LAAVHH280 pKa = 6.49RR281 pKa = 11.84RR282 pKa = 11.84FRR284 pKa = 11.84NVVEE288 pKa = 5.0FKK290 pKa = 10.68DD291 pKa = 4.88GYY293 pKa = 9.17ITTHH297 pKa = 6.45LASNYY302 pKa = 9.09FNVGATSLTPGLTTPLPWTVTDD324 pKa = 4.09RR325 pKa = 11.84PVTPTQPYY333 pKa = 8.61MSPEE337 pKa = 4.02LTDD340 pKa = 3.46SQLFATMDD348 pKa = 3.25
MM1 pKa = 7.33SVHH4 pKa = 7.06GNRR7 pKa = 11.84AWCFTLNNPTDD18 pKa = 3.84EE19 pKa = 4.69EE20 pKa = 4.41KK21 pKa = 11.15LLLPTSPEE29 pKa = 4.16SLPLWPRR36 pKa = 11.84CTALVYY42 pKa = 10.43QLEE45 pKa = 4.37EE46 pKa = 4.59GEE48 pKa = 4.55NGTPHH53 pKa = 5.5IQGYY57 pKa = 10.64ARR59 pKa = 11.84FDD61 pKa = 3.6RR62 pKa = 11.84QRR64 pKa = 11.84TLATVKK70 pKa = 10.02TILARR75 pKa = 11.84AHH77 pKa = 6.24WEE79 pKa = 3.7PAKK82 pKa = 10.92GSIQANLDD90 pKa = 3.43YY91 pKa = 10.87CGKK94 pKa = 9.58QLGRR98 pKa = 11.84LSEE101 pKa = 3.9PVIIGFGGLRR111 pKa = 11.84PGASGPSKK119 pKa = 9.99PHH121 pKa = 6.56LKK123 pKa = 10.02RR124 pKa = 11.84AQFVDD129 pKa = 6.11LIASSPDD136 pKa = 3.13VSIPEE141 pKa = 4.63LLDD144 pKa = 3.28QGALEE149 pKa = 4.36VLATQPNLLGTLRR162 pKa = 11.84GFLFQDD168 pKa = 3.76LRR170 pKa = 11.84RR171 pKa = 11.84DD172 pKa = 3.69GVIVDD177 pKa = 4.7LYY179 pKa = 11.35YY180 pKa = 11.39GLTGTGKK187 pKa = 10.18SRR189 pKa = 11.84LAFDD193 pKa = 4.69EE194 pKa = 4.39YY195 pKa = 10.65PGAFRR200 pKa = 11.84KK201 pKa = 8.41TSGSWWDD208 pKa = 3.48SYY210 pKa = 11.65AGEE213 pKa = 4.32LSVVLDD219 pKa = 4.46DD220 pKa = 5.71FDD222 pKa = 5.04DD223 pKa = 4.33AFMPIGDD230 pKa = 4.98LLRR233 pKa = 11.84TLDD236 pKa = 4.09RR237 pKa = 11.84YY238 pKa = 9.12PLRR241 pKa = 11.84MPVKK245 pKa = 10.28GGFIQLIANHH255 pKa = 6.76FIITSNHH262 pKa = 6.22LPSEE266 pKa = 4.88WYY268 pKa = 9.87PNAPPCRR275 pKa = 11.84LAAVHH280 pKa = 6.49RR281 pKa = 11.84RR282 pKa = 11.84FRR284 pKa = 11.84NVVEE288 pKa = 5.0FKK290 pKa = 10.68DD291 pKa = 4.88GYY293 pKa = 9.17ITTHH297 pKa = 6.45LASNYY302 pKa = 9.09FNVGATSLTPGLTTPLPWTVTDD324 pKa = 4.09RR325 pKa = 11.84PVTPTQPYY333 pKa = 8.61MSPEE337 pKa = 4.02LTDD340 pKa = 3.46SQLFATMDD348 pKa = 3.25
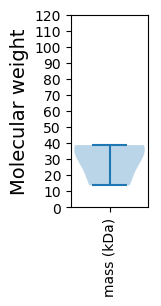
Molecular weight: 38.69 kDa
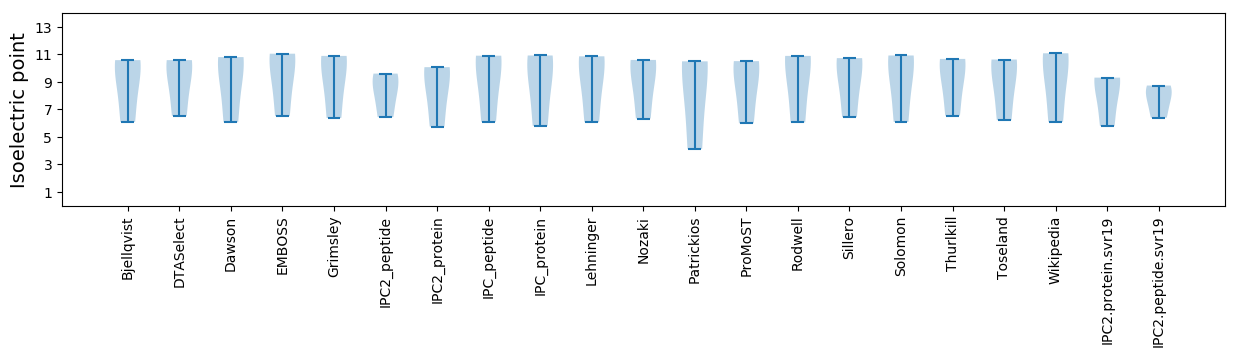
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTW0|A0A140CTW0_9VIRU Uncharacterized protein OS=Pacific flying fox faeces associated circular DNA virus-13 OX=1796008 PE=4 SV=1
MM1 pKa = 7.54PRR3 pKa = 11.84FSRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 8.92RR11 pKa = 11.84RR12 pKa = 11.84PYY14 pKa = 9.16RR15 pKa = 11.84GRR17 pKa = 11.84STRR20 pKa = 11.84SRR22 pKa = 11.84FRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84LRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84FTGRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.84RR37 pKa = 11.84YY38 pKa = 8.47FVKK41 pKa = 10.41KK42 pKa = 9.71QLVPGSSVARR52 pKa = 11.84IVMNTPIRR60 pKa = 11.84ITLGTVADD68 pKa = 4.13SRR70 pKa = 11.84VGGAWIANSLEE81 pKa = 4.13GPDD84 pKa = 5.18FISSTPGLARR94 pKa = 11.84GQYY97 pKa = 10.38SIMRR101 pKa = 11.84IAAGPSEE108 pKa = 4.67TIPIGTAGATMYY120 pKa = 10.54RR121 pKa = 11.84QGHH124 pKa = 5.95RR125 pKa = 11.84DD126 pKa = 2.84IAYY129 pKa = 8.8VYY131 pKa = 10.5SSNFMHH137 pKa = 7.61LVRR140 pKa = 11.84FYY142 pKa = 11.19RR143 pKa = 11.84NYY145 pKa = 10.57RR146 pKa = 11.84VVKK149 pKa = 7.84TTLTLSLAYY158 pKa = 10.09DD159 pKa = 3.66NRR161 pKa = 11.84PNSIDD166 pKa = 3.26QTSGMSRR173 pKa = 11.84FASVTLFATDD183 pKa = 5.26DD184 pKa = 3.58IDD186 pKa = 6.47SIDD189 pKa = 3.67TDD191 pKa = 4.38GDD193 pKa = 3.96INDD196 pKa = 3.19KK197 pKa = 10.13WIRR200 pKa = 11.84PYY202 pKa = 11.02AYY204 pKa = 9.62GHH206 pKa = 5.84RR207 pKa = 11.84HH208 pKa = 5.19VKK210 pKa = 9.36TKK212 pKa = 9.43TLHH215 pKa = 6.2NGHH218 pKa = 7.15ISPSKK223 pKa = 10.3ISMSTSAYY231 pKa = 10.36RR232 pKa = 11.84LVKK235 pKa = 10.57DD236 pKa = 3.5RR237 pKa = 11.84TTLTSADD244 pKa = 3.8YY245 pKa = 10.61VGSITPVEE253 pKa = 4.39PQPPIYY259 pKa = 9.99PGLNSPEE266 pKa = 3.92KK267 pKa = 9.72KK268 pKa = 10.4VYY270 pKa = 9.62IGFVISPYY278 pKa = 10.53GVPTGGWPASTEE290 pKa = 4.0IVSGYY295 pKa = 8.81LTVTQKK301 pKa = 10.42VQFFNPHH308 pKa = 5.93DD309 pKa = 3.57TADD312 pKa = 3.66FYY314 pKa = 11.91GPVV317 pKa = 3.15
MM1 pKa = 7.54PRR3 pKa = 11.84FSRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84YY10 pKa = 8.92RR11 pKa = 11.84RR12 pKa = 11.84PYY14 pKa = 9.16RR15 pKa = 11.84GRR17 pKa = 11.84STRR20 pKa = 11.84SRR22 pKa = 11.84FRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84LRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84FTGRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.84RR37 pKa = 11.84YY38 pKa = 8.47FVKK41 pKa = 10.41KK42 pKa = 9.71QLVPGSSVARR52 pKa = 11.84IVMNTPIRR60 pKa = 11.84ITLGTVADD68 pKa = 4.13SRR70 pKa = 11.84VGGAWIANSLEE81 pKa = 4.13GPDD84 pKa = 5.18FISSTPGLARR94 pKa = 11.84GQYY97 pKa = 10.38SIMRR101 pKa = 11.84IAAGPSEE108 pKa = 4.67TIPIGTAGATMYY120 pKa = 10.54RR121 pKa = 11.84QGHH124 pKa = 5.95RR125 pKa = 11.84DD126 pKa = 2.84IAYY129 pKa = 8.8VYY131 pKa = 10.5SSNFMHH137 pKa = 7.61LVRR140 pKa = 11.84FYY142 pKa = 11.19RR143 pKa = 11.84NYY145 pKa = 10.57RR146 pKa = 11.84VVKK149 pKa = 7.84TTLTLSLAYY158 pKa = 10.09DD159 pKa = 3.66NRR161 pKa = 11.84PNSIDD166 pKa = 3.26QTSGMSRR173 pKa = 11.84FASVTLFATDD183 pKa = 5.26DD184 pKa = 3.58IDD186 pKa = 6.47SIDD189 pKa = 3.67TDD191 pKa = 4.38GDD193 pKa = 3.96INDD196 pKa = 3.19KK197 pKa = 10.13WIRR200 pKa = 11.84PYY202 pKa = 11.02AYY204 pKa = 9.62GHH206 pKa = 5.84RR207 pKa = 11.84HH208 pKa = 5.19VKK210 pKa = 9.36TKK212 pKa = 9.43TLHH215 pKa = 6.2NGHH218 pKa = 7.15ISPSKK223 pKa = 10.3ISMSTSAYY231 pKa = 10.36RR232 pKa = 11.84LVKK235 pKa = 10.57DD236 pKa = 3.5RR237 pKa = 11.84TTLTSADD244 pKa = 3.8YY245 pKa = 10.61VGSITPVEE253 pKa = 4.39PQPPIYY259 pKa = 9.99PGLNSPEE266 pKa = 3.92KK267 pKa = 9.72KK268 pKa = 10.4VYY270 pKa = 9.62IGFVISPYY278 pKa = 10.53GVPTGGWPASTEE290 pKa = 4.0IVSGYY295 pKa = 8.81LTVTQKK301 pKa = 10.42VQFFNPHH308 pKa = 5.93DD309 pKa = 3.57TADD312 pKa = 3.66FYY314 pKa = 11.91GPVV317 pKa = 3.15
Molecular weight: 35.81 kDa
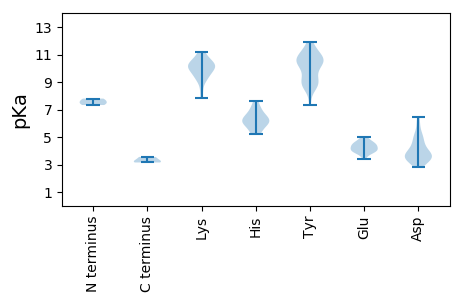
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
776 |
111 |
348 |
258.7 |
29.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.572 ± 0.511 | 0.515 ± 0.346 |
5.284 ± 0.504 | 3.351 ± 0.916 |
4.124 ± 0.383 | 7.216 ± 1.133 |
2.32 ± 0.099 | 4.768 ± 1.26 |
3.866 ± 0.877 | 9.278 ± 2.275 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.933 ± 0.29 | 3.866 ± 0.841 |
6.701 ± 1.507 | 3.479 ± 0.994 |
9.021 ± 1.474 | 7.088 ± 1.289 |
7.99 ± 0.516 | 5.67 ± 0.712 |
2.062 ± 0.887 | 4.897 ± 0.794 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
