
Prochlorococcus marinus (strain SARG / CCMP1375 / SS120)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Prochloraceae; Prochlorococcus; Prochlorococcus marinus; Prochlorococcus marinus subsp. marinus
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
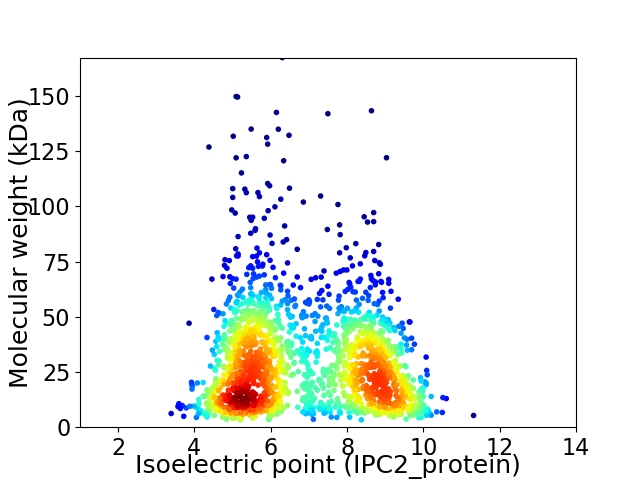
Virtual 2D-PAGE plot for 1881 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
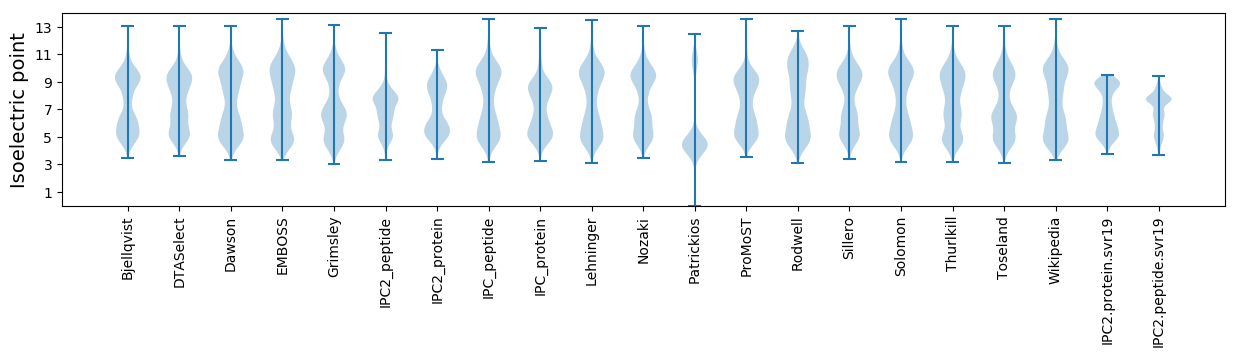
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q7VCK5|UPPP_PROMA Undecaprenyl-diphosphatase OS=Prochlorococcus marinus (strain SARG / CCMP1375 / SS120) OX=167539 GN=uppP PE=3 SV=1
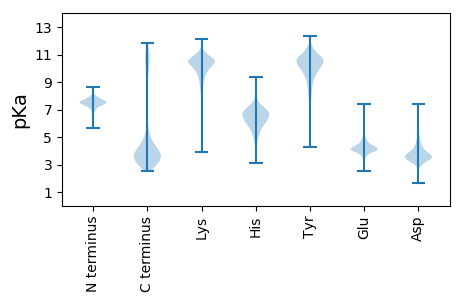
MM1 pKa = 7.62KK2 pKa = 10.48LFRR5 pKa = 11.84QLLVAPAALGLLAPLAANAAEE26 pKa = 4.18VNIKK30 pKa = 10.36DD31 pKa = 3.69VASYY35 pKa = 11.62ADD37 pKa = 4.12LNAEE41 pKa = 3.98VTTTQFSDD49 pKa = 3.75VVPGDD54 pKa = 3.31WAYY57 pKa = 8.91TALQNLSEE65 pKa = 4.8SYY67 pKa = 10.92GCVDD71 pKa = 3.74NAYY74 pKa = 7.76TQNLKK79 pKa = 10.5SGQALTRR86 pKa = 11.84YY87 pKa = 7.92EE88 pKa = 3.79AAALVNACLDD98 pKa = 3.53GGIASAEE105 pKa = 4.19VTPDD109 pKa = 3.07AARR112 pKa = 11.84LANEE116 pKa = 5.14FGTEE120 pKa = 3.7MAILKK125 pKa = 10.21GRR127 pKa = 11.84VDD129 pKa = 3.35GLEE132 pKa = 4.03YY133 pKa = 10.33KK134 pKa = 10.72VKK136 pKa = 10.1EE137 pKa = 3.98LSAGQFSSSTKK148 pKa = 10.17LSGSAVFTTGAIDD161 pKa = 4.06GMSLFDD167 pKa = 5.4DD168 pKa = 4.92PYY170 pKa = 10.25KK171 pKa = 11.09GKK173 pKa = 8.0TAFEE177 pKa = 3.87YY178 pKa = 10.43RR179 pKa = 11.84YY180 pKa = 10.54GIDD183 pKa = 4.41LNTSFNGTDD192 pKa = 3.26GLYY195 pKa = 10.74AGIEE199 pKa = 3.97QGNQDD204 pKa = 4.93DD205 pKa = 5.85LVLQDD210 pKa = 4.78AVTDD214 pKa = 3.66STLTVTSLFYY224 pKa = 10.8NFQVGEE230 pKa = 4.17FDD232 pKa = 3.76VTAGPLFDD240 pKa = 3.78QDD242 pKa = 4.63DD243 pKa = 4.72VISATTSIASEE254 pKa = 4.24SFRR257 pKa = 11.84LSSMPWGAVATTGAGAAIAYY277 pKa = 10.59SNDD280 pKa = 3.12SGFNASLSSVSSDD293 pKa = 3.62AADD296 pKa = 3.34ASKK299 pKa = 11.1GIWTDD304 pKa = 3.21GGADD308 pKa = 3.43IYY310 pKa = 10.28TASLGYY316 pKa = 10.79DD317 pKa = 2.88SDD319 pKa = 4.88YY320 pKa = 11.5YY321 pKa = 11.33GGGLIYY327 pKa = 10.55TDD329 pKa = 5.37DD330 pKa = 4.15DD331 pKa = 4.04TDD333 pKa = 3.38TSFGGGIYY341 pKa = 8.92MRR343 pKa = 11.84PDD345 pKa = 4.02GFPTISVAYY354 pKa = 7.22DD355 pKa = 3.5TKK357 pKa = 11.36DD358 pKa = 3.13PAASGTKK365 pKa = 9.94SSSHH369 pKa = 6.21LLIGVDD375 pKa = 3.69HH376 pKa = 6.65EE377 pKa = 4.81VGPGTASAAYY387 pKa = 8.22TNHH390 pKa = 6.63DD391 pKa = 3.57QMGVTGDD398 pKa = 3.43SFEE401 pKa = 4.22IYY403 pKa = 10.46YY404 pKa = 10.18NYY406 pKa = 9.66PVSDD410 pKa = 4.01GLSLQAGVFWEE421 pKa = 4.31DD422 pKa = 2.71RR423 pKa = 11.84HH424 pKa = 5.29VVKK427 pKa = 10.89NGSWTTQDD435 pKa = 2.7ATGYY439 pKa = 10.11LVEE442 pKa = 4.76TNFSFF447 pKa = 4.81
MM1 pKa = 7.62KK2 pKa = 10.48LFRR5 pKa = 11.84QLLVAPAALGLLAPLAANAAEE26 pKa = 4.18VNIKK30 pKa = 10.36DD31 pKa = 3.69VASYY35 pKa = 11.62ADD37 pKa = 4.12LNAEE41 pKa = 3.98VTTTQFSDD49 pKa = 3.75VVPGDD54 pKa = 3.31WAYY57 pKa = 8.91TALQNLSEE65 pKa = 4.8SYY67 pKa = 10.92GCVDD71 pKa = 3.74NAYY74 pKa = 7.76TQNLKK79 pKa = 10.5SGQALTRR86 pKa = 11.84YY87 pKa = 7.92EE88 pKa = 3.79AAALVNACLDD98 pKa = 3.53GGIASAEE105 pKa = 4.19VTPDD109 pKa = 3.07AARR112 pKa = 11.84LANEE116 pKa = 5.14FGTEE120 pKa = 3.7MAILKK125 pKa = 10.21GRR127 pKa = 11.84VDD129 pKa = 3.35GLEE132 pKa = 4.03YY133 pKa = 10.33KK134 pKa = 10.72VKK136 pKa = 10.1EE137 pKa = 3.98LSAGQFSSSTKK148 pKa = 10.17LSGSAVFTTGAIDD161 pKa = 4.06GMSLFDD167 pKa = 5.4DD168 pKa = 4.92PYY170 pKa = 10.25KK171 pKa = 11.09GKK173 pKa = 8.0TAFEE177 pKa = 3.87YY178 pKa = 10.43RR179 pKa = 11.84YY180 pKa = 10.54GIDD183 pKa = 4.41LNTSFNGTDD192 pKa = 3.26GLYY195 pKa = 10.74AGIEE199 pKa = 3.97QGNQDD204 pKa = 4.93DD205 pKa = 5.85LVLQDD210 pKa = 4.78AVTDD214 pKa = 3.66STLTVTSLFYY224 pKa = 10.8NFQVGEE230 pKa = 4.17FDD232 pKa = 3.76VTAGPLFDD240 pKa = 3.78QDD242 pKa = 4.63DD243 pKa = 4.72VISATTSIASEE254 pKa = 4.24SFRR257 pKa = 11.84LSSMPWGAVATTGAGAAIAYY277 pKa = 10.59SNDD280 pKa = 3.12SGFNASLSSVSSDD293 pKa = 3.62AADD296 pKa = 3.34ASKK299 pKa = 11.1GIWTDD304 pKa = 3.21GGADD308 pKa = 3.43IYY310 pKa = 10.28TASLGYY316 pKa = 10.79DD317 pKa = 2.88SDD319 pKa = 4.88YY320 pKa = 11.5YY321 pKa = 11.33GGGLIYY327 pKa = 10.55TDD329 pKa = 5.37DD330 pKa = 4.15DD331 pKa = 4.04TDD333 pKa = 3.38TSFGGGIYY341 pKa = 8.92MRR343 pKa = 11.84PDD345 pKa = 4.02GFPTISVAYY354 pKa = 7.22DD355 pKa = 3.5TKK357 pKa = 11.36DD358 pKa = 3.13PAASGTKK365 pKa = 9.94SSSHH369 pKa = 6.21LLIGVDD375 pKa = 3.69HH376 pKa = 6.65EE377 pKa = 4.81VGPGTASAAYY387 pKa = 8.22TNHH390 pKa = 6.63DD391 pKa = 3.57QMGVTGDD398 pKa = 3.43SFEE401 pKa = 4.22IYY403 pKa = 10.46YY404 pKa = 10.18NYY406 pKa = 9.66PVSDD410 pKa = 4.01GLSLQAGVFWEE421 pKa = 4.31DD422 pKa = 2.71RR423 pKa = 11.84HH424 pKa = 5.29VVKK427 pKa = 10.89NGSWTTQDD435 pKa = 2.7ATGYY439 pKa = 10.11LVEE442 pKa = 4.76TNFSFF447 pKa = 4.81
Molecular weight: 47.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q7VB24|Q7VB24_PROMA Uncharacterized protein OS=Prochlorococcus marinus (strain SARG / CCMP1375 / SS120) OX=167539 GN=Pro_1276 PE=4 SV=1
MM1 pKa = 7.41TKK3 pKa = 9.05RR4 pKa = 11.84TFGGTSRR11 pKa = 11.84KK12 pKa = 9.13RR13 pKa = 11.84KK14 pKa = 8.17RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84THH26 pKa = 6.87TGRR29 pKa = 11.84SVIRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.33KK38 pKa = 8.85GRR40 pKa = 11.84SRR42 pKa = 11.84IAVV45 pKa = 3.34
MM1 pKa = 7.41TKK3 pKa = 9.05RR4 pKa = 11.84TFGGTSRR11 pKa = 11.84KK12 pKa = 9.13RR13 pKa = 11.84KK14 pKa = 8.17RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84THH26 pKa = 6.87TGRR29 pKa = 11.84SVIRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.33KK38 pKa = 8.85GRR40 pKa = 11.84SRR42 pKa = 11.84IAVV45 pKa = 3.34
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
516670 |
30 |
1524 |
274.7 |
30.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.904 ± 0.06 | 1.221 ± 0.02 |
4.94 ± 0.04 | 6.433 ± 0.051 |
4.068 ± 0.039 | 6.879 ± 0.057 |
1.815 ± 0.031 | 7.818 ± 0.052 |
6.603 ± 0.057 | 11.523 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.949 ± 0.02 | 4.976 ± 0.044 |
4.252 ± 0.03 | 3.666 ± 0.031 |
4.93 ± 0.046 | 7.719 ± 0.045 |
4.695 ± 0.042 | 5.702 ± 0.05 |
1.461 ± 0.03 | 2.446 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
