
Capybara microvirus Cap1_SP_70
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
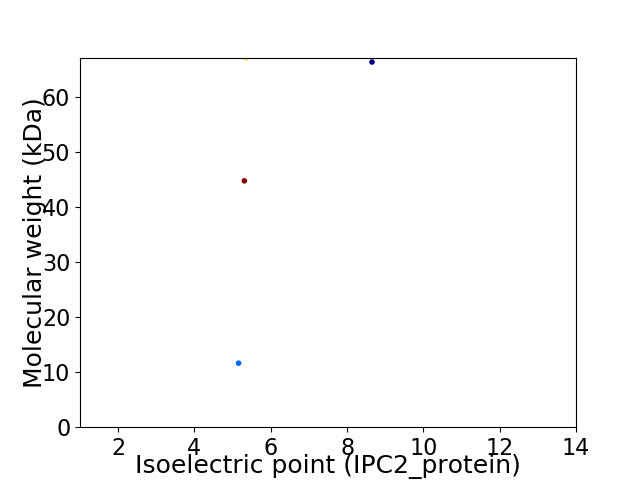
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
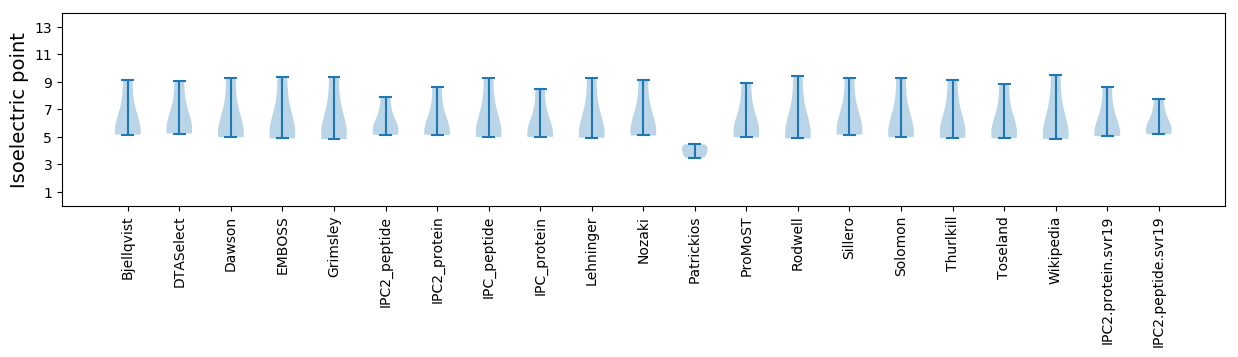
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W3Z1|A0A4P8W3Z1_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_70 OX=2584789 PE=4 SV=1
MM1 pKa = 7.76RR2 pKa = 11.84VTKK5 pKa = 10.31KK6 pKa = 10.49LYY8 pKa = 10.18KK9 pKa = 9.02PCEE12 pKa = 3.73YY13 pKa = 10.49SPRR16 pKa = 11.84SLHH19 pKa = 7.49FITNPDD25 pKa = 3.64NPDD28 pKa = 3.2VLIPEE33 pKa = 4.52CTEE36 pKa = 4.01SDD38 pKa = 3.18IKK40 pKa = 10.96FYY42 pKa = 11.13DD43 pKa = 3.58LSLVSLQNAGIDD55 pKa = 3.65VQKK58 pKa = 11.17FNLKK62 pKa = 8.99TSAPSRR68 pKa = 11.84LEE70 pKa = 3.76KK71 pKa = 10.67EE72 pKa = 4.06MSVMQMGEE80 pKa = 4.25IISKK84 pKa = 10.39HH85 pKa = 5.9FDD87 pKa = 3.22TEE89 pKa = 4.43TLDD92 pKa = 4.61KK93 pKa = 10.98PIDD96 pKa = 3.84EE97 pKa = 4.93QKK99 pKa = 9.85TEE101 pKa = 3.75
MM1 pKa = 7.76RR2 pKa = 11.84VTKK5 pKa = 10.31KK6 pKa = 10.49LYY8 pKa = 10.18KK9 pKa = 9.02PCEE12 pKa = 3.73YY13 pKa = 10.49SPRR16 pKa = 11.84SLHH19 pKa = 7.49FITNPDD25 pKa = 3.64NPDD28 pKa = 3.2VLIPEE33 pKa = 4.52CTEE36 pKa = 4.01SDD38 pKa = 3.18IKK40 pKa = 10.96FYY42 pKa = 11.13DD43 pKa = 3.58LSLVSLQNAGIDD55 pKa = 3.65VQKK58 pKa = 11.17FNLKK62 pKa = 8.99TSAPSRR68 pKa = 11.84LEE70 pKa = 3.76KK71 pKa = 10.67EE72 pKa = 4.06MSVMQMGEE80 pKa = 4.25IISKK84 pKa = 10.39HH85 pKa = 5.9FDD87 pKa = 3.22TEE89 pKa = 4.43TLDD92 pKa = 4.61KK93 pKa = 10.98PIDD96 pKa = 3.84EE97 pKa = 4.93QKK99 pKa = 9.85TEE101 pKa = 3.75
Molecular weight: 11.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4Q9|A0A4P8W4Q9_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_70 OX=2584789 PE=3 SV=1
MM1 pKa = 7.59EE2 pKa = 4.35SAFYY6 pKa = 10.6KK7 pKa = 10.32IFNPKK12 pKa = 9.32KK13 pKa = 9.36CFDD16 pKa = 3.25MRR18 pKa = 11.84YY19 pKa = 10.12DD20 pKa = 3.28KK21 pKa = 11.13DD22 pKa = 3.78YY23 pKa = 11.43FILSKK28 pKa = 10.99SNSYY32 pKa = 11.34VNMRR36 pKa = 11.84QKK38 pKa = 10.32QLSMEE43 pKa = 3.47IRR45 pKa = 11.84LYY47 pKa = 11.38YY48 pKa = 9.96EE49 pKa = 4.52FEE51 pKa = 4.32YY52 pKa = 9.33CQKK55 pKa = 10.17MNGRR59 pKa = 11.84VYY61 pKa = 10.9FLTFTYY67 pKa = 10.9NNNNIPYY74 pKa = 9.59FLGEE78 pKa = 4.13PCPDD82 pKa = 3.38KK83 pKa = 11.25KK84 pKa = 11.13DD85 pKa = 3.37MQRR88 pKa = 11.84FLNHH92 pKa = 5.76SRR94 pKa = 11.84FARR97 pKa = 11.84NLEE100 pKa = 4.08KK101 pKa = 10.28IYY103 pKa = 10.42GYY105 pKa = 10.32KK106 pKa = 10.28FKK108 pKa = 10.92YY109 pKa = 10.13FCATEE114 pKa = 4.28LGDD117 pKa = 3.94GKK119 pKa = 10.58GKK121 pKa = 10.51RR122 pKa = 11.84GIGKK126 pKa = 8.6NPHH129 pKa = 5.0FHH131 pKa = 6.93VIFFLYY137 pKa = 7.34PTSEE141 pKa = 4.3VKK143 pKa = 10.91SMSDD147 pKa = 3.11EE148 pKa = 4.05QFNILAKK155 pKa = 9.95TYY157 pKa = 8.89WQGLNDD163 pKa = 4.04NGTLKK168 pKa = 10.61DD169 pKa = 3.98YY170 pKa = 11.28KK171 pKa = 10.16SANYY175 pKa = 10.1GHH177 pKa = 6.95VSTSDD182 pKa = 2.67KK183 pKa = 10.76GSRR186 pKa = 11.84VNDD189 pKa = 3.49FRR191 pKa = 11.84AIGYY195 pKa = 5.02TAKK198 pKa = 10.68YY199 pKa = 8.41VLKK202 pKa = 10.49DD203 pKa = 2.85IYY205 pKa = 9.79TQQRR209 pKa = 11.84DD210 pKa = 3.7KK211 pKa = 11.2FFKK214 pKa = 10.25HH215 pKa = 6.05HH216 pKa = 5.59LAEE219 pKa = 4.59YY220 pKa = 9.94ISNEE224 pKa = 3.05ISKK227 pKa = 11.02YY228 pKa = 9.78IDD230 pKa = 3.62AGHH233 pKa = 6.9EE234 pKa = 3.89DD235 pKa = 3.51FDD237 pKa = 5.12FYY239 pKa = 11.4KK240 pKa = 10.84SPEE243 pKa = 4.06YY244 pKa = 11.04YY245 pKa = 8.53IDD247 pKa = 3.81KK248 pKa = 10.86YY249 pKa = 11.16GLHH252 pKa = 6.12KK253 pKa = 9.55TQDD256 pKa = 3.43PYY258 pKa = 11.93GWFHH262 pKa = 5.34QCEE265 pKa = 4.62VEE267 pKa = 3.95WLRR270 pKa = 11.84PILKK274 pKa = 10.12LRR276 pKa = 11.84FQTYY280 pKa = 9.38YY281 pKa = 10.92EE282 pKa = 4.34LLSEE286 pKa = 4.23YY287 pKa = 10.55KK288 pKa = 10.2NRR290 pKa = 11.84YY291 pKa = 4.97STKK294 pKa = 10.52PMFSHH299 pKa = 7.01NLGLYY304 pKa = 10.29ALDD307 pKa = 4.1FVTPLGKK314 pKa = 10.15VKK316 pKa = 10.65YY317 pKa = 9.78YY318 pKa = 10.83SNNNICYY325 pKa = 7.98VTLPLYY331 pKa = 10.56LYY333 pKa = 10.52RR334 pKa = 11.84KK335 pKa = 8.07FFCTVEE341 pKa = 3.8KK342 pKa = 10.48SQYY345 pKa = 11.07NFTINKK351 pKa = 8.54NGKK354 pKa = 9.33KK355 pKa = 9.46IYY357 pKa = 10.25NNIYY361 pKa = 9.0VHH363 pKa = 6.11TKK365 pKa = 9.82QGIEE369 pKa = 3.56LLYY372 pKa = 10.76RR373 pKa = 11.84QFLYY377 pKa = 10.98KK378 pKa = 10.25LDD380 pKa = 3.95LSQNVLSALIPTLQNSKK397 pKa = 10.76LVDD400 pKa = 3.43EE401 pKa = 5.96FINFQKK407 pKa = 10.8EE408 pKa = 3.45KK409 pKa = 10.27GIYY412 pKa = 9.5YY413 pKa = 7.38EE414 pKa = 4.49TSTSTVIKK422 pKa = 10.26KK423 pKa = 8.97LQSYY427 pKa = 6.96EE428 pKa = 3.81FRR430 pKa = 11.84HH431 pKa = 6.79SYY433 pKa = 10.31FLYY436 pKa = 9.36NYY438 pKa = 9.21LYY440 pKa = 10.34KK441 pKa = 10.71GRR443 pKa = 11.84YY444 pKa = 7.63FPKK447 pKa = 10.22TNTNKK452 pKa = 8.9LTYY455 pKa = 10.54NEE457 pKa = 3.96INNYY461 pKa = 9.75YY462 pKa = 10.62SVGNLDD468 pKa = 3.46IYY470 pKa = 10.54TDD472 pKa = 3.99FVRR475 pKa = 11.84FHH477 pKa = 7.01TYY479 pKa = 10.41NEE481 pKa = 4.38CVTTKK486 pKa = 10.07QRR488 pKa = 11.84QSDD491 pKa = 3.38IFEE494 pKa = 4.76SYY496 pKa = 11.37DD497 pKa = 3.3VFPPFAQNLHH507 pKa = 6.11IYY509 pKa = 9.92CWIDD513 pKa = 3.47DD514 pKa = 4.04FQHH517 pKa = 6.99FILSQSDD524 pKa = 3.64EE525 pKa = 3.9RR526 pKa = 11.84SKK528 pKa = 11.43AIFYY532 pKa = 9.46EE533 pKa = 4.3KK534 pKa = 10.52ARR536 pKa = 11.84CRR538 pKa = 11.84RR539 pKa = 11.84NLKK542 pKa = 9.35PLKK545 pKa = 10.35KK546 pKa = 9.64YY547 pKa = 10.94LCVV550 pKa = 3.39
MM1 pKa = 7.59EE2 pKa = 4.35SAFYY6 pKa = 10.6KK7 pKa = 10.32IFNPKK12 pKa = 9.32KK13 pKa = 9.36CFDD16 pKa = 3.25MRR18 pKa = 11.84YY19 pKa = 10.12DD20 pKa = 3.28KK21 pKa = 11.13DD22 pKa = 3.78YY23 pKa = 11.43FILSKK28 pKa = 10.99SNSYY32 pKa = 11.34VNMRR36 pKa = 11.84QKK38 pKa = 10.32QLSMEE43 pKa = 3.47IRR45 pKa = 11.84LYY47 pKa = 11.38YY48 pKa = 9.96EE49 pKa = 4.52FEE51 pKa = 4.32YY52 pKa = 9.33CQKK55 pKa = 10.17MNGRR59 pKa = 11.84VYY61 pKa = 10.9FLTFTYY67 pKa = 10.9NNNNIPYY74 pKa = 9.59FLGEE78 pKa = 4.13PCPDD82 pKa = 3.38KK83 pKa = 11.25KK84 pKa = 11.13DD85 pKa = 3.37MQRR88 pKa = 11.84FLNHH92 pKa = 5.76SRR94 pKa = 11.84FARR97 pKa = 11.84NLEE100 pKa = 4.08KK101 pKa = 10.28IYY103 pKa = 10.42GYY105 pKa = 10.32KK106 pKa = 10.28FKK108 pKa = 10.92YY109 pKa = 10.13FCATEE114 pKa = 4.28LGDD117 pKa = 3.94GKK119 pKa = 10.58GKK121 pKa = 10.51RR122 pKa = 11.84GIGKK126 pKa = 8.6NPHH129 pKa = 5.0FHH131 pKa = 6.93VIFFLYY137 pKa = 7.34PTSEE141 pKa = 4.3VKK143 pKa = 10.91SMSDD147 pKa = 3.11EE148 pKa = 4.05QFNILAKK155 pKa = 9.95TYY157 pKa = 8.89WQGLNDD163 pKa = 4.04NGTLKK168 pKa = 10.61DD169 pKa = 3.98YY170 pKa = 11.28KK171 pKa = 10.16SANYY175 pKa = 10.1GHH177 pKa = 6.95VSTSDD182 pKa = 2.67KK183 pKa = 10.76GSRR186 pKa = 11.84VNDD189 pKa = 3.49FRR191 pKa = 11.84AIGYY195 pKa = 5.02TAKK198 pKa = 10.68YY199 pKa = 8.41VLKK202 pKa = 10.49DD203 pKa = 2.85IYY205 pKa = 9.79TQQRR209 pKa = 11.84DD210 pKa = 3.7KK211 pKa = 11.2FFKK214 pKa = 10.25HH215 pKa = 6.05HH216 pKa = 5.59LAEE219 pKa = 4.59YY220 pKa = 9.94ISNEE224 pKa = 3.05ISKK227 pKa = 11.02YY228 pKa = 9.78IDD230 pKa = 3.62AGHH233 pKa = 6.9EE234 pKa = 3.89DD235 pKa = 3.51FDD237 pKa = 5.12FYY239 pKa = 11.4KK240 pKa = 10.84SPEE243 pKa = 4.06YY244 pKa = 11.04YY245 pKa = 8.53IDD247 pKa = 3.81KK248 pKa = 10.86YY249 pKa = 11.16GLHH252 pKa = 6.12KK253 pKa = 9.55TQDD256 pKa = 3.43PYY258 pKa = 11.93GWFHH262 pKa = 5.34QCEE265 pKa = 4.62VEE267 pKa = 3.95WLRR270 pKa = 11.84PILKK274 pKa = 10.12LRR276 pKa = 11.84FQTYY280 pKa = 9.38YY281 pKa = 10.92EE282 pKa = 4.34LLSEE286 pKa = 4.23YY287 pKa = 10.55KK288 pKa = 10.2NRR290 pKa = 11.84YY291 pKa = 4.97STKK294 pKa = 10.52PMFSHH299 pKa = 7.01NLGLYY304 pKa = 10.29ALDD307 pKa = 4.1FVTPLGKK314 pKa = 10.15VKK316 pKa = 10.65YY317 pKa = 9.78YY318 pKa = 10.83SNNNICYY325 pKa = 7.98VTLPLYY331 pKa = 10.56LYY333 pKa = 10.52RR334 pKa = 11.84KK335 pKa = 8.07FFCTVEE341 pKa = 3.8KK342 pKa = 10.48SQYY345 pKa = 11.07NFTINKK351 pKa = 8.54NGKK354 pKa = 9.33KK355 pKa = 9.46IYY357 pKa = 10.25NNIYY361 pKa = 9.0VHH363 pKa = 6.11TKK365 pKa = 9.82QGIEE369 pKa = 3.56LLYY372 pKa = 10.76RR373 pKa = 11.84QFLYY377 pKa = 10.98KK378 pKa = 10.25LDD380 pKa = 3.95LSQNVLSALIPTLQNSKK397 pKa = 10.76LVDD400 pKa = 3.43EE401 pKa = 5.96FINFQKK407 pKa = 10.8EE408 pKa = 3.45KK409 pKa = 10.27GIYY412 pKa = 9.5YY413 pKa = 7.38EE414 pKa = 4.49TSTSTVIKK422 pKa = 10.26KK423 pKa = 8.97LQSYY427 pKa = 6.96EE428 pKa = 3.81FRR430 pKa = 11.84HH431 pKa = 6.79SYY433 pKa = 10.31FLYY436 pKa = 9.36NYY438 pKa = 9.21LYY440 pKa = 10.34KK441 pKa = 10.71GRR443 pKa = 11.84YY444 pKa = 7.63FPKK447 pKa = 10.22TNTNKK452 pKa = 8.9LTYY455 pKa = 10.54NEE457 pKa = 3.96INNYY461 pKa = 9.75YY462 pKa = 10.62SVGNLDD468 pKa = 3.46IYY470 pKa = 10.54TDD472 pKa = 3.99FVRR475 pKa = 11.84FHH477 pKa = 7.01TYY479 pKa = 10.41NEE481 pKa = 4.38CVTTKK486 pKa = 10.07QRR488 pKa = 11.84QSDD491 pKa = 3.38IFEE494 pKa = 4.76SYY496 pKa = 11.37DD497 pKa = 3.3VFPPFAQNLHH507 pKa = 6.11IYY509 pKa = 9.92CWIDD513 pKa = 3.47DD514 pKa = 4.04FQHH517 pKa = 6.99FILSQSDD524 pKa = 3.64EE525 pKa = 3.9RR526 pKa = 11.84SKK528 pKa = 11.43AIFYY532 pKa = 9.46EE533 pKa = 4.3KK534 pKa = 10.52ARR536 pKa = 11.84CRR538 pKa = 11.84RR539 pKa = 11.84NLKK542 pKa = 9.35PLKK545 pKa = 10.35KK546 pKa = 9.64YY547 pKa = 10.94LCVV550 pKa = 3.39
Molecular weight: 66.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1646 |
101 |
586 |
411.5 |
47.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.739 ± 1.781 | 1.094 ± 0.375 |
5.893 ± 0.693 | 5.772 ± 1.042 |
6.075 ± 0.793 | 5.832 ± 0.99 |
1.944 ± 0.344 | 5.225 ± 0.329 |
7.23 ± 1.151 | 8.688 ± 0.164 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.187 ± 0.366 | 6.865 ± 0.53 |
3.949 ± 0.584 | 4.496 ± 0.286 |
4.374 ± 0.233 | 7.412 ± 0.497 |
5.711 ± 0.345 | 5.164 ± 0.79 |
0.972 ± 0.207 | 6.379 ± 1.785 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
