
Bat coronavirus CDPHE15/USA/2006
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Colacovirus; Bat coronavirus CDPHE15
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
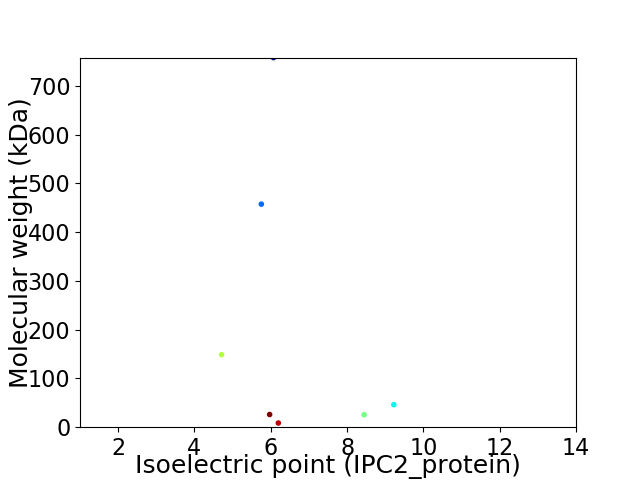
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5Z1D1|S5Z1D1_9ALPC 3C-like proteinase OS=Bat coronavirus CDPHE15/USA/2006 OX=1384461 GN=PP1a PE=3 SV=1
MM1 pKa = 7.52KK2 pKa = 10.53LIILLCIGFAYY13 pKa = 10.4ANQRR17 pKa = 11.84NVGCSEE23 pKa = 3.91GGYY26 pKa = 10.12LDD28 pKa = 3.73PPKK31 pKa = 10.39RR32 pKa = 11.84LQLGLDD38 pKa = 3.27NVTALVPGALPLPGLWNCTVTSNFIYY64 pKa = 10.41GPTPRR69 pKa = 11.84PIGLFIMYY77 pKa = 8.79YY78 pKa = 10.79ASAQVAQFGISSTLRR93 pKa = 11.84RR94 pKa = 11.84SLSPYY99 pKa = 10.35SLYY102 pKa = 10.6FLNYY106 pKa = 8.94GNGRR110 pKa = 11.84RR111 pKa = 11.84HH112 pKa = 7.63IIRR115 pKa = 11.84ICKK118 pKa = 9.0WDD120 pKa = 3.97TDD122 pKa = 4.13KK123 pKa = 11.07MLQDD127 pKa = 3.58SDD129 pKa = 3.43ATTGYY134 pKa = 11.11DD135 pKa = 3.25CIANVRR141 pKa = 11.84INDD144 pKa = 3.39VVLEE148 pKa = 4.11HH149 pKa = 7.07GGRR152 pKa = 11.84QIYY155 pKa = 9.24GLSWSGNVVTLHH167 pKa = 4.97MLKK170 pKa = 10.37GIRR173 pKa = 11.84HH174 pKa = 5.01ITVPGAQYY182 pKa = 10.06WDD184 pKa = 3.92IIRR187 pKa = 11.84FSCGHH192 pKa = 6.81KK193 pKa = 10.23DD194 pKa = 3.44SCGHH198 pKa = 4.71QVVYY202 pKa = 8.99STNVFNVTVLNGFITNYY219 pKa = 9.91QVLPDD224 pKa = 3.71SGEE227 pKa = 3.9FSDD230 pKa = 5.28NLFTVGDD237 pKa = 4.53DD238 pKa = 3.85GSIPPSFGFNNWFVLSNSSSIISGTVVSNQPLRR271 pKa = 11.84LTCLWPIPSSTGALATIYY289 pKa = 10.79FNGTNGAQCNGFDD302 pKa = 4.3SNAPFDD308 pKa = 5.01AIRR311 pKa = 11.84FNLNGTLSGHH321 pKa = 5.69NFVSGFVLHH330 pKa = 7.0AANGATLGFSCTNSTDD346 pKa = 3.35APYY349 pKa = 10.78LRR351 pKa = 11.84QIPFGIGDD359 pKa = 3.76TPYY362 pKa = 10.11YY363 pKa = 10.47CYY365 pKa = 11.19LNVTTDD371 pKa = 3.03INSTMSFVGALPLNLRR387 pKa = 11.84EE388 pKa = 4.61IVIASNGDD396 pKa = 3.24VYY398 pKa = 11.19MNGYY402 pKa = 10.17RR403 pKa = 11.84YY404 pKa = 9.08FAAGDD409 pKa = 3.79LSSVDD414 pKa = 3.53VEE416 pKa = 4.87LPSQQVFGSTFWTIAFTVFEE436 pKa = 4.47TVLLEE441 pKa = 4.39VDD443 pKa = 3.48GTSINRR449 pKa = 11.84MLYY452 pKa = 10.15CDD454 pKa = 3.72NPLNRR459 pKa = 11.84VKK461 pKa = 10.65CSHH464 pKa = 5.61TQFDD468 pKa = 4.9LVDD471 pKa = 3.79GFYY474 pKa = 10.76PLTDD478 pKa = 2.53VDD480 pKa = 4.64LAVKK484 pKa = 10.14PFTFVTLPTFADD496 pKa = 3.34HH497 pKa = 6.83SFVYY501 pKa = 10.33FNFSLMFDD509 pKa = 4.32DD510 pKa = 5.11LNEE513 pKa = 4.18DD514 pKa = 3.64FRR516 pKa = 11.84LQSFNLTINGQLSYY530 pKa = 10.5CVQSRR535 pKa = 11.84QFTTSGSVRR544 pKa = 11.84TNTNHH549 pKa = 5.92QFGFYY554 pKa = 6.84TQRR557 pKa = 11.84AASNGCPFTIDD568 pKa = 3.24TLNNYY573 pKa = 7.78LTFGRR578 pKa = 11.84ICFSFGEE585 pKa = 4.41SGAGCGVDD593 pKa = 4.15VMVEE597 pKa = 3.81SQYY600 pKa = 12.11NMFKK604 pKa = 9.36VTTIFVSYY612 pKa = 10.94SEE614 pKa = 4.22GDD616 pKa = 3.62IIAGMPKK623 pKa = 9.47PSEE626 pKa = 4.44GIRR629 pKa = 11.84DD630 pKa = 3.47VSKK633 pKa = 10.75IHH635 pKa = 7.31LDD637 pKa = 3.37VCSTYY642 pKa = 10.67SIYY645 pKa = 11.25GHH647 pKa = 6.51TGDD650 pKa = 5.26GIIRR654 pKa = 11.84LTNDD658 pKa = 2.99TLLGGLYY665 pKa = 8.39YY666 pKa = 10.15TSPGGALLGFKK677 pKa = 10.4NVTTGEE683 pKa = 4.25IYY685 pKa = 10.72SVTPCSLTQQVAVVSDD701 pKa = 4.18EE702 pKa = 3.93IVGVVSSSANVSDD715 pKa = 3.66QFSFTYY721 pKa = 8.02TTVTEE726 pKa = 3.96QFYY729 pKa = 11.38YY730 pKa = 8.1MTDD733 pKa = 3.51GNSSCSEE740 pKa = 3.81PVLTYY745 pKa = 11.04SSLGVCKK752 pKa = 10.68DD753 pKa = 2.92GALRR757 pKa = 11.84ILQPRR762 pKa = 11.84EE763 pKa = 3.99DD764 pKa = 3.92TTQPTPIVSGVISIATNFTLSVVTEE789 pKa = 4.24YY790 pKa = 10.55IQLANTPISVDD801 pKa = 2.97CAMYY805 pKa = 10.43VCNGNPRR812 pKa = 11.84CNMLLAQYY820 pKa = 10.93SSACQTIEE828 pKa = 4.23NALQLSAKK836 pKa = 10.03LEE838 pKa = 4.54SIEE841 pKa = 4.04VTNMLTVSEE850 pKa = 5.24DD851 pKa = 3.22NLQIANISTFNGGGYY866 pKa = 10.45NFTNLMGTTYY876 pKa = 9.11STKK879 pKa = 10.65SVVEE883 pKa = 4.96DD884 pKa = 3.16ILFDD888 pKa = 3.77KK889 pKa = 11.09VVTSGLGTVDD899 pKa = 4.39ADD901 pKa = 4.53YY902 pKa = 10.68KK903 pKa = 10.81ACSNGLSIADD913 pKa = 4.03LVCAQYY919 pKa = 10.49YY920 pKa = 8.27QGVMVLPGVVDD931 pKa = 3.56AAKK934 pKa = 10.54LHH936 pKa = 6.26MYY938 pKa = 9.71SASLMGGMALGGVTAAAALPFSYY961 pKa = 10.43AVQARR966 pKa = 11.84LNYY969 pKa = 9.72VALQTDD975 pKa = 3.94VLQRR979 pKa = 11.84NQQILAEE986 pKa = 4.36SFNNAIGNITNAFASVNDD1004 pKa = 5.22AISQTAEE1011 pKa = 3.87GLSTVAEE1018 pKa = 4.35ALSKK1022 pKa = 10.66VQDD1025 pKa = 3.63VVNNQGMALNHH1036 pKa = 6.33LTLQLQNNFQAISSSIADD1054 pKa = 3.56IYY1056 pKa = 11.12RR1057 pKa = 11.84RR1058 pKa = 11.84LDD1060 pKa = 3.25QLTADD1065 pKa = 3.8AQVDD1069 pKa = 3.65RR1070 pKa = 11.84LINGRR1075 pKa = 11.84LAALNAFVSQTLTKK1089 pKa = 9.99YY1090 pKa = 10.58SQVQASRR1097 pKa = 11.84SLAKK1101 pKa = 10.31QKK1103 pKa = 10.63INEE1106 pKa = 4.48CVLSQSPRR1114 pKa = 11.84YY1115 pKa = 9.08GFCGDD1120 pKa = 3.65GGRR1123 pKa = 11.84HH1124 pKa = 5.01VFTVTQAAPQGILFLHH1140 pKa = 5.98TVLRR1144 pKa = 11.84PTGSVNVTAAAGICVDD1160 pKa = 3.52GAGYY1164 pKa = 11.02ALNQPGLVLIYY1175 pKa = 10.35QDD1177 pKa = 3.53GTYY1180 pKa = 10.64LITPRR1185 pKa = 11.84VMFEE1189 pKa = 3.61PRR1191 pKa = 11.84QPQISDD1197 pKa = 3.33FVRR1200 pKa = 11.84IEE1202 pKa = 4.28GCDD1205 pKa = 3.25VEE1207 pKa = 4.56YY1208 pKa = 11.02FNVTGEE1214 pKa = 4.21SLPDD1218 pKa = 3.53IFPDD1222 pKa = 5.29FIDD1225 pKa = 3.66VNKK1228 pKa = 9.33TLEE1231 pKa = 5.24DD1232 pKa = 3.61ILSQLQNNTGPKK1244 pKa = 9.75FDD1246 pKa = 4.18IDD1248 pKa = 3.92IFNATYY1254 pKa = 11.05LNLSSEE1260 pKa = 4.22IADD1263 pKa = 3.29LEE1265 pKa = 4.21MRR1267 pKa = 11.84SEE1269 pKa = 4.2SLHH1272 pKa = 5.37NTTEE1276 pKa = 3.98EE1277 pKa = 4.09LKK1279 pKa = 10.96RR1280 pKa = 11.84LIDD1283 pKa = 4.57NINSTLVDD1291 pKa = 4.3LEE1293 pKa = 3.91WLNRR1297 pKa = 11.84VEE1299 pKa = 5.97TYY1301 pKa = 10.41IKK1303 pKa = 9.23WPWWVWLLIAIALIFTVSLLLFCCIATGCCGCCGCCASCLTGCCKK1348 pKa = 10.37GPRR1351 pKa = 11.84LQPYY1355 pKa = 8.15EE1356 pKa = 4.76AIEE1359 pKa = 4.2KK1360 pKa = 9.87VHH1362 pKa = 5.3VQQ1364 pKa = 3.06
MM1 pKa = 7.52KK2 pKa = 10.53LIILLCIGFAYY13 pKa = 10.4ANQRR17 pKa = 11.84NVGCSEE23 pKa = 3.91GGYY26 pKa = 10.12LDD28 pKa = 3.73PPKK31 pKa = 10.39RR32 pKa = 11.84LQLGLDD38 pKa = 3.27NVTALVPGALPLPGLWNCTVTSNFIYY64 pKa = 10.41GPTPRR69 pKa = 11.84PIGLFIMYY77 pKa = 8.79YY78 pKa = 10.79ASAQVAQFGISSTLRR93 pKa = 11.84RR94 pKa = 11.84SLSPYY99 pKa = 10.35SLYY102 pKa = 10.6FLNYY106 pKa = 8.94GNGRR110 pKa = 11.84RR111 pKa = 11.84HH112 pKa = 7.63IIRR115 pKa = 11.84ICKK118 pKa = 9.0WDD120 pKa = 3.97TDD122 pKa = 4.13KK123 pKa = 11.07MLQDD127 pKa = 3.58SDD129 pKa = 3.43ATTGYY134 pKa = 11.11DD135 pKa = 3.25CIANVRR141 pKa = 11.84INDD144 pKa = 3.39VVLEE148 pKa = 4.11HH149 pKa = 7.07GGRR152 pKa = 11.84QIYY155 pKa = 9.24GLSWSGNVVTLHH167 pKa = 4.97MLKK170 pKa = 10.37GIRR173 pKa = 11.84HH174 pKa = 5.01ITVPGAQYY182 pKa = 10.06WDD184 pKa = 3.92IIRR187 pKa = 11.84FSCGHH192 pKa = 6.81KK193 pKa = 10.23DD194 pKa = 3.44SCGHH198 pKa = 4.71QVVYY202 pKa = 8.99STNVFNVTVLNGFITNYY219 pKa = 9.91QVLPDD224 pKa = 3.71SGEE227 pKa = 3.9FSDD230 pKa = 5.28NLFTVGDD237 pKa = 4.53DD238 pKa = 3.85GSIPPSFGFNNWFVLSNSSSIISGTVVSNQPLRR271 pKa = 11.84LTCLWPIPSSTGALATIYY289 pKa = 10.79FNGTNGAQCNGFDD302 pKa = 4.3SNAPFDD308 pKa = 5.01AIRR311 pKa = 11.84FNLNGTLSGHH321 pKa = 5.69NFVSGFVLHH330 pKa = 7.0AANGATLGFSCTNSTDD346 pKa = 3.35APYY349 pKa = 10.78LRR351 pKa = 11.84QIPFGIGDD359 pKa = 3.76TPYY362 pKa = 10.11YY363 pKa = 10.47CYY365 pKa = 11.19LNVTTDD371 pKa = 3.03INSTMSFVGALPLNLRR387 pKa = 11.84EE388 pKa = 4.61IVIASNGDD396 pKa = 3.24VYY398 pKa = 11.19MNGYY402 pKa = 10.17RR403 pKa = 11.84YY404 pKa = 9.08FAAGDD409 pKa = 3.79LSSVDD414 pKa = 3.53VEE416 pKa = 4.87LPSQQVFGSTFWTIAFTVFEE436 pKa = 4.47TVLLEE441 pKa = 4.39VDD443 pKa = 3.48GTSINRR449 pKa = 11.84MLYY452 pKa = 10.15CDD454 pKa = 3.72NPLNRR459 pKa = 11.84VKK461 pKa = 10.65CSHH464 pKa = 5.61TQFDD468 pKa = 4.9LVDD471 pKa = 3.79GFYY474 pKa = 10.76PLTDD478 pKa = 2.53VDD480 pKa = 4.64LAVKK484 pKa = 10.14PFTFVTLPTFADD496 pKa = 3.34HH497 pKa = 6.83SFVYY501 pKa = 10.33FNFSLMFDD509 pKa = 4.32DD510 pKa = 5.11LNEE513 pKa = 4.18DD514 pKa = 3.64FRR516 pKa = 11.84LQSFNLTINGQLSYY530 pKa = 10.5CVQSRR535 pKa = 11.84QFTTSGSVRR544 pKa = 11.84TNTNHH549 pKa = 5.92QFGFYY554 pKa = 6.84TQRR557 pKa = 11.84AASNGCPFTIDD568 pKa = 3.24TLNNYY573 pKa = 7.78LTFGRR578 pKa = 11.84ICFSFGEE585 pKa = 4.41SGAGCGVDD593 pKa = 4.15VMVEE597 pKa = 3.81SQYY600 pKa = 12.11NMFKK604 pKa = 9.36VTTIFVSYY612 pKa = 10.94SEE614 pKa = 4.22GDD616 pKa = 3.62IIAGMPKK623 pKa = 9.47PSEE626 pKa = 4.44GIRR629 pKa = 11.84DD630 pKa = 3.47VSKK633 pKa = 10.75IHH635 pKa = 7.31LDD637 pKa = 3.37VCSTYY642 pKa = 10.67SIYY645 pKa = 11.25GHH647 pKa = 6.51TGDD650 pKa = 5.26GIIRR654 pKa = 11.84LTNDD658 pKa = 2.99TLLGGLYY665 pKa = 8.39YY666 pKa = 10.15TSPGGALLGFKK677 pKa = 10.4NVTTGEE683 pKa = 4.25IYY685 pKa = 10.72SVTPCSLTQQVAVVSDD701 pKa = 4.18EE702 pKa = 3.93IVGVVSSSANVSDD715 pKa = 3.66QFSFTYY721 pKa = 8.02TTVTEE726 pKa = 3.96QFYY729 pKa = 11.38YY730 pKa = 8.1MTDD733 pKa = 3.51GNSSCSEE740 pKa = 3.81PVLTYY745 pKa = 11.04SSLGVCKK752 pKa = 10.68DD753 pKa = 2.92GALRR757 pKa = 11.84ILQPRR762 pKa = 11.84EE763 pKa = 3.99DD764 pKa = 3.92TTQPTPIVSGVISIATNFTLSVVTEE789 pKa = 4.24YY790 pKa = 10.55IQLANTPISVDD801 pKa = 2.97CAMYY805 pKa = 10.43VCNGNPRR812 pKa = 11.84CNMLLAQYY820 pKa = 10.93SSACQTIEE828 pKa = 4.23NALQLSAKK836 pKa = 10.03LEE838 pKa = 4.54SIEE841 pKa = 4.04VTNMLTVSEE850 pKa = 5.24DD851 pKa = 3.22NLQIANISTFNGGGYY866 pKa = 10.45NFTNLMGTTYY876 pKa = 9.11STKK879 pKa = 10.65SVVEE883 pKa = 4.96DD884 pKa = 3.16ILFDD888 pKa = 3.77KK889 pKa = 11.09VVTSGLGTVDD899 pKa = 4.39ADD901 pKa = 4.53YY902 pKa = 10.68KK903 pKa = 10.81ACSNGLSIADD913 pKa = 4.03LVCAQYY919 pKa = 10.49YY920 pKa = 8.27QGVMVLPGVVDD931 pKa = 3.56AAKK934 pKa = 10.54LHH936 pKa = 6.26MYY938 pKa = 9.71SASLMGGMALGGVTAAAALPFSYY961 pKa = 10.43AVQARR966 pKa = 11.84LNYY969 pKa = 9.72VALQTDD975 pKa = 3.94VLQRR979 pKa = 11.84NQQILAEE986 pKa = 4.36SFNNAIGNITNAFASVNDD1004 pKa = 5.22AISQTAEE1011 pKa = 3.87GLSTVAEE1018 pKa = 4.35ALSKK1022 pKa = 10.66VQDD1025 pKa = 3.63VVNNQGMALNHH1036 pKa = 6.33LTLQLQNNFQAISSSIADD1054 pKa = 3.56IYY1056 pKa = 11.12RR1057 pKa = 11.84RR1058 pKa = 11.84LDD1060 pKa = 3.25QLTADD1065 pKa = 3.8AQVDD1069 pKa = 3.65RR1070 pKa = 11.84LINGRR1075 pKa = 11.84LAALNAFVSQTLTKK1089 pKa = 9.99YY1090 pKa = 10.58SQVQASRR1097 pKa = 11.84SLAKK1101 pKa = 10.31QKK1103 pKa = 10.63INEE1106 pKa = 4.48CVLSQSPRR1114 pKa = 11.84YY1115 pKa = 9.08GFCGDD1120 pKa = 3.65GGRR1123 pKa = 11.84HH1124 pKa = 5.01VFTVTQAAPQGILFLHH1140 pKa = 5.98TVLRR1144 pKa = 11.84PTGSVNVTAAAGICVDD1160 pKa = 3.52GAGYY1164 pKa = 11.02ALNQPGLVLIYY1175 pKa = 10.35QDD1177 pKa = 3.53GTYY1180 pKa = 10.64LITPRR1185 pKa = 11.84VMFEE1189 pKa = 3.61PRR1191 pKa = 11.84QPQISDD1197 pKa = 3.33FVRR1200 pKa = 11.84IEE1202 pKa = 4.28GCDD1205 pKa = 3.25VEE1207 pKa = 4.56YY1208 pKa = 11.02FNVTGEE1214 pKa = 4.21SLPDD1218 pKa = 3.53IFPDD1222 pKa = 5.29FIDD1225 pKa = 3.66VNKK1228 pKa = 9.33TLEE1231 pKa = 5.24DD1232 pKa = 3.61ILSQLQNNTGPKK1244 pKa = 9.75FDD1246 pKa = 4.18IDD1248 pKa = 3.92IFNATYY1254 pKa = 11.05LNLSSEE1260 pKa = 4.22IADD1263 pKa = 3.29LEE1265 pKa = 4.21MRR1267 pKa = 11.84SEE1269 pKa = 4.2SLHH1272 pKa = 5.37NTTEE1276 pKa = 3.98EE1277 pKa = 4.09LKK1279 pKa = 10.96RR1280 pKa = 11.84LIDD1283 pKa = 4.57NINSTLVDD1291 pKa = 4.3LEE1293 pKa = 3.91WLNRR1297 pKa = 11.84VEE1299 pKa = 5.97TYY1301 pKa = 10.41IKK1303 pKa = 9.23WPWWVWLLIAIALIFTVSLLLFCCIATGCCGCCGCCASCLTGCCKK1348 pKa = 10.37GPRR1351 pKa = 11.84LQPYY1355 pKa = 8.15EE1356 pKa = 4.76AIEE1359 pKa = 4.2KK1360 pKa = 9.87VHH1362 pKa = 5.3VQQ1364 pKa = 3.06
Molecular weight: 148.89 kDa
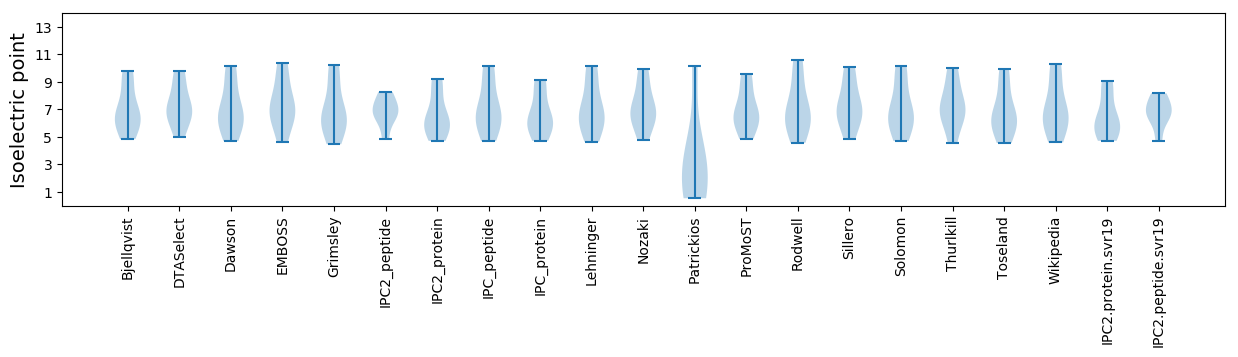
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5YGS4|S5YGS4_9ALPC Envelope small membrane protein OS=Bat coronavirus CDPHE15/USA/2006 OX=1384461 GN=E PE=3 SV=1
MM1 pKa = 7.64ASVKK5 pKa = 9.64FAKK8 pKa = 10.16NNRR11 pKa = 11.84RR12 pKa = 11.84GRR14 pKa = 11.84MEE16 pKa = 3.78YY17 pKa = 10.41SYY19 pKa = 10.63FAPLIINSDD28 pKa = 3.55QPIWKK33 pKa = 9.29VLPNNAVPVGKK44 pKa = 10.24GGKK47 pKa = 9.4DD48 pKa = 3.23EE49 pKa = 5.24QIGYY53 pKa = 9.25WNEE56 pKa = 3.57QPRR59 pKa = 11.84WRR61 pKa = 11.84MRR63 pKa = 11.84KK64 pKa = 7.43GQRR67 pKa = 11.84VDD69 pKa = 3.85VASKK73 pKa = 8.05WHH75 pKa = 6.62FYY77 pKa = 10.96YY78 pKa = 10.84LGTGPHH84 pKa = 6.25SEE86 pKa = 3.69EE87 pKa = 4.24QYY89 pKa = 10.56RR90 pKa = 11.84KK91 pKa = 10.32RR92 pKa = 11.84IDD94 pKa = 3.15GVYY97 pKa = 8.29WVAVNGAKK105 pKa = 9.64TSPTGLGTRR114 pKa = 11.84SAKK117 pKa = 9.37TKK119 pKa = 10.01PLEE122 pKa = 4.15LKK124 pKa = 10.67FNTKK128 pKa = 9.27IPKK131 pKa = 9.19EE132 pKa = 4.13VEE134 pKa = 4.1FVEE137 pKa = 4.64PTSPGSSRR145 pKa = 11.84ANSRR149 pKa = 11.84SQSRR153 pKa = 11.84GGSKK157 pKa = 10.13SRR159 pKa = 11.84NNSPRR164 pKa = 11.84RR165 pKa = 11.84GNQSKK170 pKa = 10.5SRR172 pKa = 11.84NSSKK176 pKa = 10.66NRR178 pKa = 11.84GGDD181 pKa = 3.15NRR183 pKa = 11.84SRR185 pKa = 11.84SSSGTRR191 pKa = 11.84DD192 pKa = 2.69QDD194 pKa = 4.04AIVAAVKK201 pKa = 10.22AALLGLGLGTDD212 pKa = 4.06SNSGASGKK220 pKa = 10.21ASKK223 pKa = 10.78ANSGTSTPKK232 pKa = 9.99PKK234 pKa = 9.88PAATPKK240 pKa = 10.66SPSTPKK246 pKa = 10.45SQLEE250 pKa = 3.89RR251 pKa = 11.84PEE253 pKa = 4.03WKK255 pKa = 9.89RR256 pKa = 11.84VPDD259 pKa = 3.54SDD261 pKa = 5.32CSVEE265 pKa = 4.07QCFGPRR271 pKa = 11.84GGFKK275 pKa = 10.84NFGSADD281 pKa = 3.43FVQYY285 pKa = 10.46GVAAKK290 pKa = 10.1GYY292 pKa = 7.96PQAAALTPTSAALLFGGNVQVQEE315 pKa = 4.42LSDD318 pKa = 4.97DD319 pKa = 3.62IEE321 pKa = 3.82ITYY324 pKa = 8.33TYY326 pKa = 10.99KK327 pKa = 8.4MTVPKK332 pKa = 10.12SDD334 pKa = 3.48KK335 pKa = 10.19NLEE338 pKa = 4.17VFLSNVNAYY347 pKa = 9.61KK348 pKa = 10.07EE349 pKa = 4.33AKK351 pKa = 8.67PQRR354 pKa = 11.84EE355 pKa = 4.15PKK357 pKa = 9.72KK358 pKa = 10.81KK359 pKa = 9.47KK360 pKa = 9.94DD361 pKa = 3.34RR362 pKa = 11.84SSRR365 pKa = 11.84PPTPAPSAPVASGNAEE381 pKa = 4.3PIYY384 pKa = 11.02ADD386 pKa = 3.74VQPQGQTEE394 pKa = 4.11PVYY397 pKa = 10.55EE398 pKa = 4.8NPDD401 pKa = 3.29QYY403 pKa = 11.47LGGVDD408 pKa = 3.5IVNEE412 pKa = 4.36VYY414 pKa = 11.11GNLPDD419 pKa = 4.58NSQSTAA425 pKa = 3.02
MM1 pKa = 7.64ASVKK5 pKa = 9.64FAKK8 pKa = 10.16NNRR11 pKa = 11.84RR12 pKa = 11.84GRR14 pKa = 11.84MEE16 pKa = 3.78YY17 pKa = 10.41SYY19 pKa = 10.63FAPLIINSDD28 pKa = 3.55QPIWKK33 pKa = 9.29VLPNNAVPVGKK44 pKa = 10.24GGKK47 pKa = 9.4DD48 pKa = 3.23EE49 pKa = 5.24QIGYY53 pKa = 9.25WNEE56 pKa = 3.57QPRR59 pKa = 11.84WRR61 pKa = 11.84MRR63 pKa = 11.84KK64 pKa = 7.43GQRR67 pKa = 11.84VDD69 pKa = 3.85VASKK73 pKa = 8.05WHH75 pKa = 6.62FYY77 pKa = 10.96YY78 pKa = 10.84LGTGPHH84 pKa = 6.25SEE86 pKa = 3.69EE87 pKa = 4.24QYY89 pKa = 10.56RR90 pKa = 11.84KK91 pKa = 10.32RR92 pKa = 11.84IDD94 pKa = 3.15GVYY97 pKa = 8.29WVAVNGAKK105 pKa = 9.64TSPTGLGTRR114 pKa = 11.84SAKK117 pKa = 9.37TKK119 pKa = 10.01PLEE122 pKa = 4.15LKK124 pKa = 10.67FNTKK128 pKa = 9.27IPKK131 pKa = 9.19EE132 pKa = 4.13VEE134 pKa = 4.1FVEE137 pKa = 4.64PTSPGSSRR145 pKa = 11.84ANSRR149 pKa = 11.84SQSRR153 pKa = 11.84GGSKK157 pKa = 10.13SRR159 pKa = 11.84NNSPRR164 pKa = 11.84RR165 pKa = 11.84GNQSKK170 pKa = 10.5SRR172 pKa = 11.84NSSKK176 pKa = 10.66NRR178 pKa = 11.84GGDD181 pKa = 3.15NRR183 pKa = 11.84SRR185 pKa = 11.84SSSGTRR191 pKa = 11.84DD192 pKa = 2.69QDD194 pKa = 4.04AIVAAVKK201 pKa = 10.22AALLGLGLGTDD212 pKa = 4.06SNSGASGKK220 pKa = 10.21ASKK223 pKa = 10.78ANSGTSTPKK232 pKa = 9.99PKK234 pKa = 9.88PAATPKK240 pKa = 10.66SPSTPKK246 pKa = 10.45SQLEE250 pKa = 3.89RR251 pKa = 11.84PEE253 pKa = 4.03WKK255 pKa = 9.89RR256 pKa = 11.84VPDD259 pKa = 3.54SDD261 pKa = 5.32CSVEE265 pKa = 4.07QCFGPRR271 pKa = 11.84GGFKK275 pKa = 10.84NFGSADD281 pKa = 3.43FVQYY285 pKa = 10.46GVAAKK290 pKa = 10.1GYY292 pKa = 7.96PQAAALTPTSAALLFGGNVQVQEE315 pKa = 4.42LSDD318 pKa = 4.97DD319 pKa = 3.62IEE321 pKa = 3.82ITYY324 pKa = 8.33TYY326 pKa = 10.99KK327 pKa = 8.4MTVPKK332 pKa = 10.12SDD334 pKa = 3.48KK335 pKa = 10.19NLEE338 pKa = 4.17VFLSNVNAYY347 pKa = 9.61KK348 pKa = 10.07EE349 pKa = 4.33AKK351 pKa = 8.67PQRR354 pKa = 11.84EE355 pKa = 4.15PKK357 pKa = 9.72KK358 pKa = 10.81KK359 pKa = 9.47KK360 pKa = 9.94DD361 pKa = 3.34RR362 pKa = 11.84SSRR365 pKa = 11.84PPTPAPSAPVASGNAEE381 pKa = 4.3PIYY384 pKa = 11.02ADD386 pKa = 3.74VQPQGQTEE394 pKa = 4.11PVYY397 pKa = 10.55EE398 pKa = 4.8NPDD401 pKa = 3.29QYY403 pKa = 11.47LGGVDD408 pKa = 3.5IVNEE412 pKa = 4.36VYY414 pKa = 11.11GNLPDD419 pKa = 4.58NSQSTAA425 pKa = 3.02
Molecular weight: 46.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13299 |
74 |
6825 |
1899.9 |
210.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.331 ± 0.266 | 3.444 ± 0.491 |
5.677 ± 0.331 | 3.812 ± 0.165 |
5.76 ± 0.297 | 7.053 ± 0.235 |
1.948 ± 0.225 | 4.579 ± 0.343 |
5.03 ± 0.549 | 8.82 ± 0.357 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.023 ± 0.192 | 5.233 ± 0.269 |
3.632 ± 0.484 | 2.918 ± 0.37 |
3.79 ± 0.275 | 7.392 ± 0.399 |
6.068 ± 0.293 | 9.873 ± 0.485 |
1.188 ± 0.139 | 4.429 ± 0.142 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
