
Hyphomicrobium facile
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Hyphomicrobiaceae; Hyphomicrobium
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
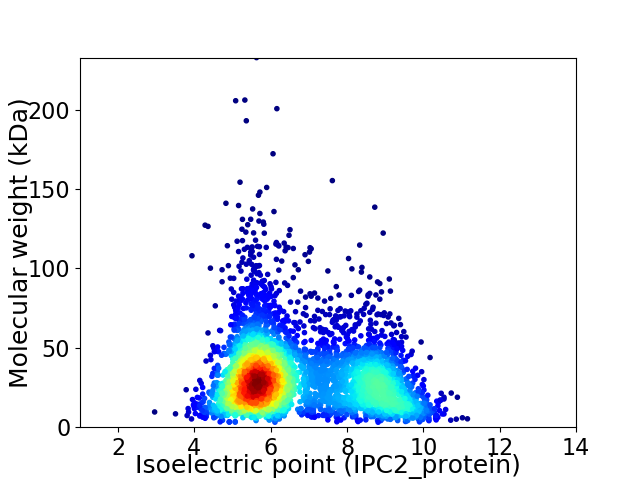
Virtual 2D-PAGE plot for 4102 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I7NBE3|A0A1I7NBE3_9RHIZ Ca-activated chloride channel family protein OS=Hyphomicrobium facile OX=51670 GN=SAMN04488557_1458 PE=4 SV=1
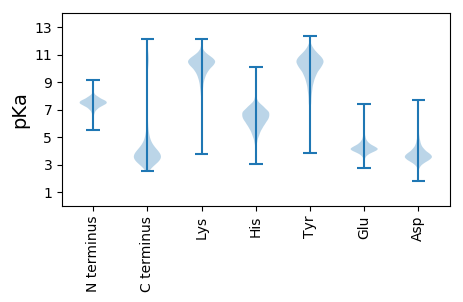
MM1 pKa = 7.43SNAIVLEE8 pKa = 4.01NEE10 pKa = 4.38KK11 pKa = 10.58PGAPQSEE18 pKa = 4.36WMLQPGEE25 pKa = 4.14ASSSIEE31 pKa = 3.85GFTTSMSTNLGSTVNFKK48 pKa = 10.87INTDD52 pKa = 3.63SNDD55 pKa = 3.23YY56 pKa = 10.85RR57 pKa = 11.84IDD59 pKa = 3.43IYY61 pKa = 11.26RR62 pKa = 11.84LGYY65 pKa = 10.15YY66 pKa = 10.35GGDD69 pKa = 3.35GATLVTSMEE78 pKa = 4.0HH79 pKa = 5.53QGAVNQPNPITDD91 pKa = 3.69PTTGNIDD98 pKa = 3.35AGNWSVTDD106 pKa = 3.17SWNIPTNATSGVYY119 pKa = 9.05IAKK122 pKa = 8.91LTRR125 pKa = 11.84LDD127 pKa = 3.6GTSGEE132 pKa = 4.23NIIPFVVTNNASTSDD147 pKa = 3.87IIFQTSDD154 pKa = 3.65STWQAYY160 pKa = 7.28NAWGGGSFYY169 pKa = 10.7TDD171 pKa = 2.96TAQNISYY178 pKa = 9.52NRR180 pKa = 11.84PIQTPAGPDD189 pKa = 3.23DD190 pKa = 4.05TTASGPWDD198 pKa = 3.48FVFGEE203 pKa = 4.62EE204 pKa = 4.69LPAIYY209 pKa = 9.99FLEE212 pKa = 4.33KK213 pKa = 10.29NGYY216 pKa = 8.72DD217 pKa = 3.22VSYY220 pKa = 11.14QSGIDD225 pKa = 3.3TATNGSLLLNHH236 pKa = 7.02KK237 pKa = 10.38AFLSVGHH244 pKa = 6.7DD245 pKa = 4.03EE246 pKa = 4.43YY247 pKa = 10.1WTAQQQNNVRR257 pKa = 11.84AARR260 pKa = 11.84DD261 pKa = 3.38AGVSLNFWSGNEE273 pKa = 4.32SYY275 pKa = 7.43WTTEE279 pKa = 4.15LLPSIDD285 pKa = 4.06GSGDD289 pKa = 3.42PNRR292 pKa = 11.84TLVTYY297 pKa = 7.8KK298 pKa = 10.33TSATGLQNPDD308 pKa = 3.83GVWTGATRR316 pKa = 11.84DD317 pKa = 3.77PAGGLIPEE325 pKa = 4.88NGLSGQMYY333 pKa = 9.91YY334 pKa = 10.94VDD336 pKa = 4.08WDD338 pKa = 3.92HH339 pKa = 6.41NTPLNPITVPYY350 pKa = 10.23KK351 pKa = 10.05YY352 pKa = 10.5AQLNIWKK359 pKa = 8.31NTEE362 pKa = 3.88VANLQPGQTYY372 pKa = 8.74TSQSDD377 pKa = 3.82YY378 pKa = 11.36LGYY381 pKa = 10.49EE382 pKa = 3.65WDD384 pKa = 3.85VNATSSEE391 pKa = 4.1VANNGFSPAGLVPLSSTTVTSDD413 pKa = 3.03AVSNAYY419 pKa = 9.58ILTGLITGTATHH431 pKa = 6.72NLTLYY436 pKa = 9.55RR437 pKa = 11.84APSGALVFAAGSIMWSWSLSSLHH460 pKa = 6.21TPGPDD465 pKa = 3.74GSPTDD470 pKa = 4.26APADD474 pKa = 3.59PAFQQAMVNLLAQEE488 pKa = 5.53GIQPQSLDD496 pKa = 3.31SSLVLASQSTDD507 pKa = 3.54FLPPTTVISTNTAAQFVAGQLVTITGTATDD537 pKa = 3.24QGGGVVAGVEE547 pKa = 4.22VSTDD551 pKa = 3.34GGNTWHH557 pKa = 6.84EE558 pKa = 4.4ANFAAAAANVSWSYY572 pKa = 9.5TWTPAATGSYY582 pKa = 9.18TILARR587 pKa = 11.84SVDD590 pKa = 3.86DD591 pKa = 4.07VANLAHH597 pKa = 7.26PLLSPNFSITDD608 pKa = 4.39FYY610 pKa = 11.7SNFAVSQGWASADD623 pKa = 3.63TLRR626 pKa = 11.84TLADD630 pKa = 3.78FNGDD634 pKa = 2.86GRR636 pKa = 11.84ADD638 pKa = 3.54FVGFGASTVYY648 pKa = 10.86ASASLGDD655 pKa = 3.46ASQYY659 pKa = 11.25NGGASFSTSLRR670 pKa = 11.84SLANDD675 pKa = 3.39FTIAQGYY682 pKa = 5.53TAANQRR688 pKa = 11.84GMDD691 pKa = 3.7FVGNLSSSPTDD702 pKa = 3.22HH703 pKa = 7.65FDD705 pKa = 3.55TLWAVGADD713 pKa = 3.56GFHH716 pKa = 6.25YY717 pKa = 10.71ASVASSGAATGSKK730 pKa = 10.16VYY732 pKa = 11.14NNFGLAQGWTQTYY745 pKa = 9.66GVDD748 pKa = 3.29VAFLSKK754 pKa = 10.49SDD756 pKa = 3.68AYY758 pKa = 10.87ASIVGFGVSGLWVGQQAFNPTATPAASYY786 pKa = 10.07IAAGSQTLGGASGWDD801 pKa = 3.34STLDD805 pKa = 3.08IQTFRR810 pKa = 11.84DD811 pKa = 3.59YY812 pKa = 10.7RR813 pKa = 11.84GNMIDD818 pKa = 4.73LNNDD822 pKa = 4.11GIADD826 pKa = 4.17FVGMGPTGLEE836 pKa = 3.72YY837 pKa = 10.33AYY839 pKa = 10.66GQFTAGATAGSQVYY853 pKa = 10.58SLGAIQKK860 pKa = 10.14GSTGTNIDD868 pKa = 3.54FGRR871 pKa = 11.84DD872 pKa = 3.57QGWDD876 pKa = 2.95TSTSDD881 pKa = 5.43RR882 pKa = 11.84ILADD886 pKa = 3.04INGDD890 pKa = 3.11GRR892 pKa = 11.84LDD894 pKa = 3.41VVAFGDD900 pKa = 3.44AGVWVSIGQQANADD914 pKa = 3.42GSGAFGAAYY923 pKa = 10.1LAMANFGTQQGWSTAVNTRR942 pKa = 11.84VLGDD946 pKa = 3.56LYY948 pKa = 11.45GNGKK952 pKa = 9.13IDD954 pKa = 3.28IVGFGADD961 pKa = 3.48YY962 pKa = 10.54TFIATPTVDD971 pKa = 2.96AATGHH976 pKa = 4.58VTFGISEE983 pKa = 4.26SLHH986 pKa = 6.91AYY988 pKa = 6.86GTNEE992 pKa = 4.52GYY994 pKa = 10.36SPSQNFRR1001 pKa = 11.84GVADD1005 pKa = 3.71VNGTGVDD1012 pKa = 4.05SIVVSSLSNTQIVSHH1027 pKa = 5.68VV1028 pKa = 3.29
MM1 pKa = 7.43SNAIVLEE8 pKa = 4.01NEE10 pKa = 4.38KK11 pKa = 10.58PGAPQSEE18 pKa = 4.36WMLQPGEE25 pKa = 4.14ASSSIEE31 pKa = 3.85GFTTSMSTNLGSTVNFKK48 pKa = 10.87INTDD52 pKa = 3.63SNDD55 pKa = 3.23YY56 pKa = 10.85RR57 pKa = 11.84IDD59 pKa = 3.43IYY61 pKa = 11.26RR62 pKa = 11.84LGYY65 pKa = 10.15YY66 pKa = 10.35GGDD69 pKa = 3.35GATLVTSMEE78 pKa = 4.0HH79 pKa = 5.53QGAVNQPNPITDD91 pKa = 3.69PTTGNIDD98 pKa = 3.35AGNWSVTDD106 pKa = 3.17SWNIPTNATSGVYY119 pKa = 9.05IAKK122 pKa = 8.91LTRR125 pKa = 11.84LDD127 pKa = 3.6GTSGEE132 pKa = 4.23NIIPFVVTNNASTSDD147 pKa = 3.87IIFQTSDD154 pKa = 3.65STWQAYY160 pKa = 7.28NAWGGGSFYY169 pKa = 10.7TDD171 pKa = 2.96TAQNISYY178 pKa = 9.52NRR180 pKa = 11.84PIQTPAGPDD189 pKa = 3.23DD190 pKa = 4.05TTASGPWDD198 pKa = 3.48FVFGEE203 pKa = 4.62EE204 pKa = 4.69LPAIYY209 pKa = 9.99FLEE212 pKa = 4.33KK213 pKa = 10.29NGYY216 pKa = 8.72DD217 pKa = 3.22VSYY220 pKa = 11.14QSGIDD225 pKa = 3.3TATNGSLLLNHH236 pKa = 7.02KK237 pKa = 10.38AFLSVGHH244 pKa = 6.7DD245 pKa = 4.03EE246 pKa = 4.43YY247 pKa = 10.1WTAQQQNNVRR257 pKa = 11.84AARR260 pKa = 11.84DD261 pKa = 3.38AGVSLNFWSGNEE273 pKa = 4.32SYY275 pKa = 7.43WTTEE279 pKa = 4.15LLPSIDD285 pKa = 4.06GSGDD289 pKa = 3.42PNRR292 pKa = 11.84TLVTYY297 pKa = 7.8KK298 pKa = 10.33TSATGLQNPDD308 pKa = 3.83GVWTGATRR316 pKa = 11.84DD317 pKa = 3.77PAGGLIPEE325 pKa = 4.88NGLSGQMYY333 pKa = 9.91YY334 pKa = 10.94VDD336 pKa = 4.08WDD338 pKa = 3.92HH339 pKa = 6.41NTPLNPITVPYY350 pKa = 10.23KK351 pKa = 10.05YY352 pKa = 10.5AQLNIWKK359 pKa = 8.31NTEE362 pKa = 3.88VANLQPGQTYY372 pKa = 8.74TSQSDD377 pKa = 3.82YY378 pKa = 11.36LGYY381 pKa = 10.49EE382 pKa = 3.65WDD384 pKa = 3.85VNATSSEE391 pKa = 4.1VANNGFSPAGLVPLSSTTVTSDD413 pKa = 3.03AVSNAYY419 pKa = 9.58ILTGLITGTATHH431 pKa = 6.72NLTLYY436 pKa = 9.55RR437 pKa = 11.84APSGALVFAAGSIMWSWSLSSLHH460 pKa = 6.21TPGPDD465 pKa = 3.74GSPTDD470 pKa = 4.26APADD474 pKa = 3.59PAFQQAMVNLLAQEE488 pKa = 5.53GIQPQSLDD496 pKa = 3.31SSLVLASQSTDD507 pKa = 3.54FLPPTTVISTNTAAQFVAGQLVTITGTATDD537 pKa = 3.24QGGGVVAGVEE547 pKa = 4.22VSTDD551 pKa = 3.34GGNTWHH557 pKa = 6.84EE558 pKa = 4.4ANFAAAAANVSWSYY572 pKa = 9.5TWTPAATGSYY582 pKa = 9.18TILARR587 pKa = 11.84SVDD590 pKa = 3.86DD591 pKa = 4.07VANLAHH597 pKa = 7.26PLLSPNFSITDD608 pKa = 4.39FYY610 pKa = 11.7SNFAVSQGWASADD623 pKa = 3.63TLRR626 pKa = 11.84TLADD630 pKa = 3.78FNGDD634 pKa = 2.86GRR636 pKa = 11.84ADD638 pKa = 3.54FVGFGASTVYY648 pKa = 10.86ASASLGDD655 pKa = 3.46ASQYY659 pKa = 11.25NGGASFSTSLRR670 pKa = 11.84SLANDD675 pKa = 3.39FTIAQGYY682 pKa = 5.53TAANQRR688 pKa = 11.84GMDD691 pKa = 3.7FVGNLSSSPTDD702 pKa = 3.22HH703 pKa = 7.65FDD705 pKa = 3.55TLWAVGADD713 pKa = 3.56GFHH716 pKa = 6.25YY717 pKa = 10.71ASVASSGAATGSKK730 pKa = 10.16VYY732 pKa = 11.14NNFGLAQGWTQTYY745 pKa = 9.66GVDD748 pKa = 3.29VAFLSKK754 pKa = 10.49SDD756 pKa = 3.68AYY758 pKa = 10.87ASIVGFGVSGLWVGQQAFNPTATPAASYY786 pKa = 10.07IAAGSQTLGGASGWDD801 pKa = 3.34STLDD805 pKa = 3.08IQTFRR810 pKa = 11.84DD811 pKa = 3.59YY812 pKa = 10.7RR813 pKa = 11.84GNMIDD818 pKa = 4.73LNNDD822 pKa = 4.11GIADD826 pKa = 4.17FVGMGPTGLEE836 pKa = 3.72YY837 pKa = 10.33AYY839 pKa = 10.66GQFTAGATAGSQVYY853 pKa = 10.58SLGAIQKK860 pKa = 10.14GSTGTNIDD868 pKa = 3.54FGRR871 pKa = 11.84DD872 pKa = 3.57QGWDD876 pKa = 2.95TSTSDD881 pKa = 5.43RR882 pKa = 11.84ILADD886 pKa = 3.04INGDD890 pKa = 3.11GRR892 pKa = 11.84LDD894 pKa = 3.41VVAFGDD900 pKa = 3.44AGVWVSIGQQANADD914 pKa = 3.42GSGAFGAAYY923 pKa = 10.1LAMANFGTQQGWSTAVNTRR942 pKa = 11.84VLGDD946 pKa = 3.56LYY948 pKa = 11.45GNGKK952 pKa = 9.13IDD954 pKa = 3.28IVGFGADD961 pKa = 3.48YY962 pKa = 10.54TFIATPTVDD971 pKa = 2.96AATGHH976 pKa = 4.58VTFGISEE983 pKa = 4.26SLHH986 pKa = 6.91AYY988 pKa = 6.86GTNEE992 pKa = 4.52GYY994 pKa = 10.36SPSQNFRR1001 pKa = 11.84GVADD1005 pKa = 3.71VNGTGVDD1012 pKa = 4.05SIVVSSLSNTQIVSHH1027 pKa = 5.68VV1028 pKa = 3.29
Molecular weight: 107.94 kDa
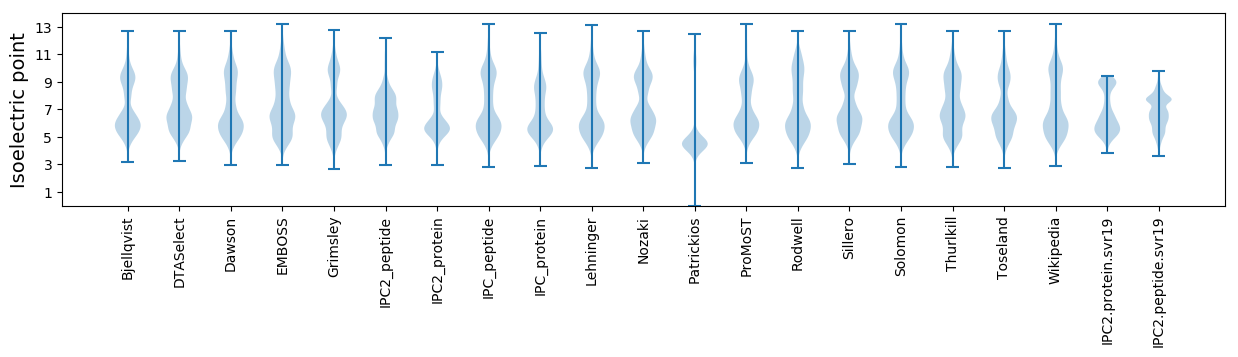
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I7NUV3|A0A1I7NUV3_9RHIZ Uncharacterized protein OS=Hyphomicrobium facile OX=51670 GN=SAMN04488557_3666 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.26RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.73GFRR19 pKa = 11.84KK20 pKa = 9.99RR21 pKa = 11.84MLTAGGQKK29 pKa = 9.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.51GRR39 pKa = 11.84KK40 pKa = 8.72RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.2QPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.26RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.73GFRR19 pKa = 11.84KK20 pKa = 9.99RR21 pKa = 11.84MLTAGGQKK29 pKa = 9.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.51GRR39 pKa = 11.84KK40 pKa = 8.72RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1277010 |
29 |
2151 |
311.3 |
33.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.01 ± 0.052 | 0.864 ± 0.014 |
5.575 ± 0.029 | 5.612 ± 0.037 |
3.813 ± 0.026 | 8.156 ± 0.037 |
1.985 ± 0.018 | 5.535 ± 0.022 |
4.096 ± 0.036 | 9.746 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.287 ± 0.016 | 2.891 ± 0.022 |
5.236 ± 0.03 | 3.049 ± 0.022 |
6.733 ± 0.042 | 6.106 ± 0.028 |
5.363 ± 0.026 | 7.304 ± 0.031 |
1.324 ± 0.016 | 2.313 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
