
[Clostridium] ultunense Esp
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Tissierellia; Tissierellales; Tissierellaceae; Schnuerera; Schnuerera ultunensis
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
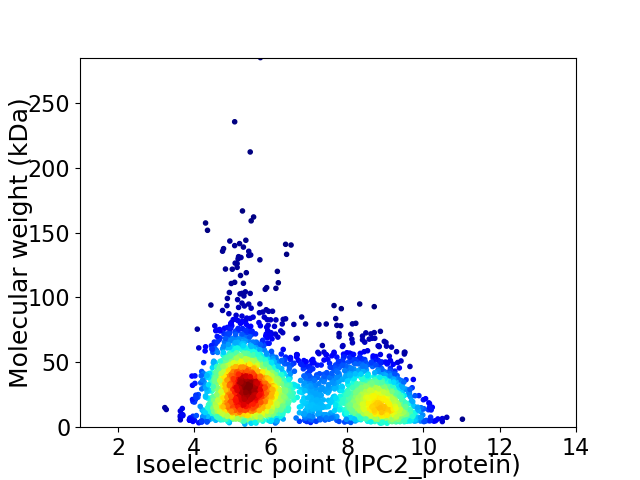
Virtual 2D-PAGE plot for 3144 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M1ZI51|M1ZI51_9FIRM Uncharacterized protein OS=[Clostridium] ultunense Esp OX=1288971 GN=CUESP1_0680 PE=4 SV=1
MM1 pKa = 7.78KK2 pKa = 10.37KK3 pKa = 10.32RR4 pKa = 11.84ILSLTLVICLLISLLVGCGQKK25 pKa = 10.86DD26 pKa = 3.41SDD28 pKa = 4.71SEE30 pKa = 4.54SDD32 pKa = 3.58TPSTGNAAEE41 pKa = 4.93DD42 pKa = 4.08GVKK45 pKa = 10.18FAVLFGTGGLGDD57 pKa = 3.5NGYY60 pKa = 10.58NDD62 pKa = 4.03EE63 pKa = 4.35VYY65 pKa = 10.17KK66 pKa = 10.05GCEE69 pKa = 3.69MAVEE73 pKa = 3.95QLDD76 pKa = 3.64ASFDD80 pKa = 3.64YY81 pKa = 11.22CEE83 pKa = 4.61PKK85 pKa = 10.55DD86 pKa = 3.63VAEE89 pKa = 5.29FEE91 pKa = 4.48TQLRR95 pKa = 11.84SYY97 pKa = 10.63ADD99 pKa = 3.2SGEE102 pKa = 3.96YY103 pKa = 10.37DD104 pKa = 3.78VIIAISTEE112 pKa = 3.99QVDD115 pKa = 3.98ALKK118 pKa = 10.1MVAEE122 pKa = 5.4EE123 pKa = 4.09YY124 pKa = 10.32QDD126 pKa = 4.36QKK128 pKa = 11.61FCMLDD133 pKa = 3.67TIVEE137 pKa = 4.62GYY139 pKa = 11.23DD140 pKa = 3.48NIHH143 pKa = 6.81SISAAYY149 pKa = 8.88PDD151 pKa = 3.48QHH153 pKa = 6.52FLSGMLAGLATQDD166 pKa = 3.44EE167 pKa = 5.18RR168 pKa = 11.84FPLSNPEE175 pKa = 3.91NVLGFCIAMDD185 pKa = 3.99TPTSRR190 pKa = 11.84GQAAGFIAGAKK201 pKa = 8.69YY202 pKa = 10.57VNPDD206 pKa = 3.14VEE208 pKa = 4.36ILTNYY213 pKa = 8.85IGAYY217 pKa = 9.61NDD219 pKa = 3.54PATAKK224 pKa = 9.96EE225 pKa = 4.2LALMMYY231 pKa = 9.93EE232 pKa = 4.19RR233 pKa = 11.84GADD236 pKa = 2.94IVSANAGSSTQGVFYY251 pKa = 10.5AAEE254 pKa = 4.03EE255 pKa = 3.98KK256 pKa = 10.43DD257 pKa = 3.46RR258 pKa = 11.84YY259 pKa = 10.78VIGTSLAMADD269 pKa = 4.56PDD271 pKa = 4.43HH272 pKa = 6.79SLCTSLKK279 pKa = 9.74KK280 pKa = 10.4VWLFVVQEE288 pKa = 4.19IEE290 pKa = 4.23SLQNGTWEE298 pKa = 4.3PGNTVMGIPEE308 pKa = 4.72GVCNYY313 pKa = 10.26DD314 pKa = 3.16IEE316 pKa = 4.55GLNVEE321 pKa = 4.11IPEE324 pKa = 5.01DD325 pKa = 3.17IVAILDD331 pKa = 3.54EE332 pKa = 4.45ARR334 pKa = 11.84EE335 pKa = 4.14LIANGDD341 pKa = 3.86LVLPTDD347 pKa = 5.08LDD349 pKa = 4.79QIDD352 pKa = 3.65EE353 pKa = 4.36WASQNQYY360 pKa = 10.92NKK362 pKa = 10.86
MM1 pKa = 7.78KK2 pKa = 10.37KK3 pKa = 10.32RR4 pKa = 11.84ILSLTLVICLLISLLVGCGQKK25 pKa = 10.86DD26 pKa = 3.41SDD28 pKa = 4.71SEE30 pKa = 4.54SDD32 pKa = 3.58TPSTGNAAEE41 pKa = 4.93DD42 pKa = 4.08GVKK45 pKa = 10.18FAVLFGTGGLGDD57 pKa = 3.5NGYY60 pKa = 10.58NDD62 pKa = 4.03EE63 pKa = 4.35VYY65 pKa = 10.17KK66 pKa = 10.05GCEE69 pKa = 3.69MAVEE73 pKa = 3.95QLDD76 pKa = 3.64ASFDD80 pKa = 3.64YY81 pKa = 11.22CEE83 pKa = 4.61PKK85 pKa = 10.55DD86 pKa = 3.63VAEE89 pKa = 5.29FEE91 pKa = 4.48TQLRR95 pKa = 11.84SYY97 pKa = 10.63ADD99 pKa = 3.2SGEE102 pKa = 3.96YY103 pKa = 10.37DD104 pKa = 3.78VIIAISTEE112 pKa = 3.99QVDD115 pKa = 3.98ALKK118 pKa = 10.1MVAEE122 pKa = 5.4EE123 pKa = 4.09YY124 pKa = 10.32QDD126 pKa = 4.36QKK128 pKa = 11.61FCMLDD133 pKa = 3.67TIVEE137 pKa = 4.62GYY139 pKa = 11.23DD140 pKa = 3.48NIHH143 pKa = 6.81SISAAYY149 pKa = 8.88PDD151 pKa = 3.48QHH153 pKa = 6.52FLSGMLAGLATQDD166 pKa = 3.44EE167 pKa = 5.18RR168 pKa = 11.84FPLSNPEE175 pKa = 3.91NVLGFCIAMDD185 pKa = 3.99TPTSRR190 pKa = 11.84GQAAGFIAGAKK201 pKa = 8.69YY202 pKa = 10.57VNPDD206 pKa = 3.14VEE208 pKa = 4.36ILTNYY213 pKa = 8.85IGAYY217 pKa = 9.61NDD219 pKa = 3.54PATAKK224 pKa = 9.96EE225 pKa = 4.2LALMMYY231 pKa = 9.93EE232 pKa = 4.19RR233 pKa = 11.84GADD236 pKa = 2.94IVSANAGSSTQGVFYY251 pKa = 10.5AAEE254 pKa = 4.03EE255 pKa = 3.98KK256 pKa = 10.43DD257 pKa = 3.46RR258 pKa = 11.84YY259 pKa = 10.78VIGTSLAMADD269 pKa = 4.56PDD271 pKa = 4.43HH272 pKa = 6.79SLCTSLKK279 pKa = 9.74KK280 pKa = 10.4VWLFVVQEE288 pKa = 4.19IEE290 pKa = 4.23SLQNGTWEE298 pKa = 4.3PGNTVMGIPEE308 pKa = 4.72GVCNYY313 pKa = 10.26DD314 pKa = 3.16IEE316 pKa = 4.55GLNVEE321 pKa = 4.11IPEE324 pKa = 5.01DD325 pKa = 3.17IVAILDD331 pKa = 3.54EE332 pKa = 4.45ARR334 pKa = 11.84EE335 pKa = 4.14LIANGDD341 pKa = 3.86LVLPTDD347 pKa = 5.08LDD349 pKa = 4.79QIDD352 pKa = 3.65EE353 pKa = 4.36WASQNQYY360 pKa = 10.92NKK362 pKa = 10.86
Molecular weight: 39.4 kDa
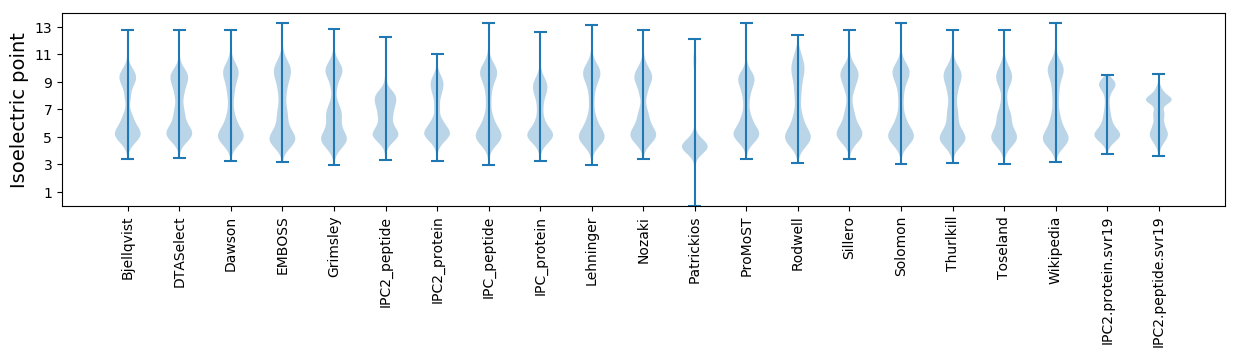
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M1ZLZ7|M1ZLZ7_9FIRM Uncharacterized protein OS=[Clostridium] ultunense Esp OX=1288971 GN=CUESP1_0681 PE=4 SV=1
MM1 pKa = 7.39NNNGFRR7 pKa = 11.84NGALLGSILGASLGMFFGTRR27 pKa = 11.84MGPMQRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84IMRR38 pKa = 11.84TAKK41 pKa = 10.02RR42 pKa = 11.84AKK44 pKa = 8.88STLLNGMNSLWGG56 pKa = 3.62
MM1 pKa = 7.39NNNGFRR7 pKa = 11.84NGALLGSILGASLGMFFGTRR27 pKa = 11.84MGPMQRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84IMRR38 pKa = 11.84TAKK41 pKa = 10.02RR42 pKa = 11.84AKK44 pKa = 8.88STLLNGMNSLWGG56 pKa = 3.62
Molecular weight: 6.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
870061 |
20 |
2452 |
276.7 |
31.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.39 ± 0.048 | 0.857 ± 0.016 |
5.658 ± 0.037 | 7.924 ± 0.053 |
4.239 ± 0.031 | 6.904 ± 0.047 |
1.485 ± 0.016 | 10.441 ± 0.054 |
8.485 ± 0.042 | 9.373 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.697 ± 0.02 | 5.645 ± 0.041 |
3.159 ± 0.027 | 2.285 ± 0.023 |
3.987 ± 0.03 | 5.683 ± 0.037 |
4.767 ± 0.029 | 6.196 ± 0.041 |
0.685 ± 0.014 | 4.138 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
