
Croton yellow vein mosaic alphasatellite
Taxonomy: Viruses; Alphasatellitidae; Geminialphasatellitinae; Clecrusatellite
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D3HTY0|D3HTY0_9VIRU Replication protein OS=Croton yellow vein mosaic alphasatellite OX=713089 GN=alpha rep PE=4 SV=1
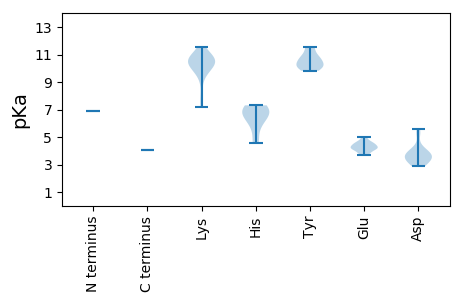
MM1 pKa = 6.91SHH3 pKa = 5.9SRR5 pKa = 11.84NWCFTIFNYY14 pKa = 9.97VLPLFTTLPEE24 pKa = 3.79WANYY28 pKa = 10.27LVFQEE33 pKa = 4.98EE34 pKa = 4.62EE35 pKa = 4.36CPTTKK40 pKa = 9.85KK41 pKa = 10.48RR42 pKa = 11.84HH43 pKa = 4.56IQGYY47 pKa = 9.82VNLKK51 pKa = 9.19RR52 pKa = 11.84NQRR55 pKa = 11.84FAFLKK60 pKa = 10.48KK61 pKa = 9.96KK62 pKa = 10.7LPDD65 pKa = 3.85GSHH68 pKa = 6.86IEE70 pKa = 4.0PCRR73 pKa = 11.84GSASSNRR80 pKa = 11.84DD81 pKa = 3.15YY82 pKa = 10.47CTKK85 pKa = 10.47DD86 pKa = 2.88ASRR89 pKa = 11.84TGGPWEE95 pKa = 4.17FGVFCEE101 pKa = 4.38TGSNKK106 pKa = 9.76RR107 pKa = 11.84KK108 pKa = 7.94TMEE111 pKa = 4.3RR112 pKa = 11.84FQEE115 pKa = 4.28DD116 pKa = 4.08PEE118 pKa = 4.17EE119 pKa = 4.61LRR121 pKa = 11.84LADD124 pKa = 3.63PKK126 pKa = 10.93LYY128 pKa = 10.25RR129 pKa = 11.84RR130 pKa = 11.84CLATKK135 pKa = 10.51VNTEE139 pKa = 3.71FSGLVLPVPDD149 pKa = 5.19RR150 pKa = 11.84PWQLVAQKK158 pKa = 10.95ALDD161 pKa = 3.56QGPDD165 pKa = 3.18DD166 pKa = 4.05RR167 pKa = 11.84TIIWVYY173 pKa = 10.5GSEE176 pKa = 4.1GNEE179 pKa = 4.89GKK181 pKa = 7.15TTWAKK186 pKa = 10.56IKK188 pKa = 10.3IQEE191 pKa = 3.75GWFYY195 pKa = 11.47SRR197 pKa = 11.84GGKK200 pKa = 9.87GEE202 pKa = 4.13NIKK205 pKa = 10.35YY206 pKa = 10.12QYY208 pKa = 10.88AEE210 pKa = 4.16HH211 pKa = 6.98LGHH214 pKa = 6.82CVFDD218 pKa = 3.66IPRR221 pKa = 11.84QVEE224 pKa = 4.24DD225 pKa = 3.31NLQYY229 pKa = 10.79TVLEE233 pKa = 4.5EE234 pKa = 3.95IKK236 pKa = 10.78DD237 pKa = 3.69RR238 pKa = 11.84LIRR241 pKa = 11.84SSKK244 pKa = 10.56YY245 pKa = 10.02EE246 pKa = 4.51PIDD249 pKa = 3.78FNCSDD254 pKa = 3.65KK255 pKa = 11.19VHH257 pKa = 6.57LVVLSNFLPCLDD269 pKa = 3.29VEE271 pKa = 4.56YY272 pKa = 10.84NNRR275 pKa = 11.84GEE277 pKa = 4.25LVKK280 pKa = 10.8KK281 pKa = 10.14PLLSRR286 pKa = 11.84DD287 pKa = 2.91RR288 pKa = 11.84VFIINIDD295 pKa = 3.63EE296 pKa = 4.59SVCGHH301 pKa = 7.3PDD303 pKa = 3.24DD304 pKa = 5.56LKK306 pKa = 11.53NFDD309 pKa = 4.28VYY311 pKa = 11.54LEE313 pKa = 4.05
MM1 pKa = 6.91SHH3 pKa = 5.9SRR5 pKa = 11.84NWCFTIFNYY14 pKa = 9.97VLPLFTTLPEE24 pKa = 3.79WANYY28 pKa = 10.27LVFQEE33 pKa = 4.98EE34 pKa = 4.62EE35 pKa = 4.36CPTTKK40 pKa = 9.85KK41 pKa = 10.48RR42 pKa = 11.84HH43 pKa = 4.56IQGYY47 pKa = 9.82VNLKK51 pKa = 9.19RR52 pKa = 11.84NQRR55 pKa = 11.84FAFLKK60 pKa = 10.48KK61 pKa = 9.96KK62 pKa = 10.7LPDD65 pKa = 3.85GSHH68 pKa = 6.86IEE70 pKa = 4.0PCRR73 pKa = 11.84GSASSNRR80 pKa = 11.84DD81 pKa = 3.15YY82 pKa = 10.47CTKK85 pKa = 10.47DD86 pKa = 2.88ASRR89 pKa = 11.84TGGPWEE95 pKa = 4.17FGVFCEE101 pKa = 4.38TGSNKK106 pKa = 9.76RR107 pKa = 11.84KK108 pKa = 7.94TMEE111 pKa = 4.3RR112 pKa = 11.84FQEE115 pKa = 4.28DD116 pKa = 4.08PEE118 pKa = 4.17EE119 pKa = 4.61LRR121 pKa = 11.84LADD124 pKa = 3.63PKK126 pKa = 10.93LYY128 pKa = 10.25RR129 pKa = 11.84RR130 pKa = 11.84CLATKK135 pKa = 10.51VNTEE139 pKa = 3.71FSGLVLPVPDD149 pKa = 5.19RR150 pKa = 11.84PWQLVAQKK158 pKa = 10.95ALDD161 pKa = 3.56QGPDD165 pKa = 3.18DD166 pKa = 4.05RR167 pKa = 11.84TIIWVYY173 pKa = 10.5GSEE176 pKa = 4.1GNEE179 pKa = 4.89GKK181 pKa = 7.15TTWAKK186 pKa = 10.56IKK188 pKa = 10.3IQEE191 pKa = 3.75GWFYY195 pKa = 11.47SRR197 pKa = 11.84GGKK200 pKa = 9.87GEE202 pKa = 4.13NIKK205 pKa = 10.35YY206 pKa = 10.12QYY208 pKa = 10.88AEE210 pKa = 4.16HH211 pKa = 6.98LGHH214 pKa = 6.82CVFDD218 pKa = 3.66IPRR221 pKa = 11.84QVEE224 pKa = 4.24DD225 pKa = 3.31NLQYY229 pKa = 10.79TVLEE233 pKa = 4.5EE234 pKa = 3.95IKK236 pKa = 10.78DD237 pKa = 3.69RR238 pKa = 11.84LIRR241 pKa = 11.84SSKK244 pKa = 10.56YY245 pKa = 10.02EE246 pKa = 4.51PIDD249 pKa = 3.78FNCSDD254 pKa = 3.65KK255 pKa = 11.19VHH257 pKa = 6.57LVVLSNFLPCLDD269 pKa = 3.29VEE271 pKa = 4.56YY272 pKa = 10.84NNRR275 pKa = 11.84GEE277 pKa = 4.25LVKK280 pKa = 10.8KK281 pKa = 10.14PLLSRR286 pKa = 11.84DD287 pKa = 2.91RR288 pKa = 11.84VFIINIDD295 pKa = 3.63EE296 pKa = 4.59SVCGHH301 pKa = 7.3PDD303 pKa = 3.24DD304 pKa = 5.56LKK306 pKa = 11.53NFDD309 pKa = 4.28VYY311 pKa = 11.54LEE313 pKa = 4.05
Molecular weight: 36.45 kDa
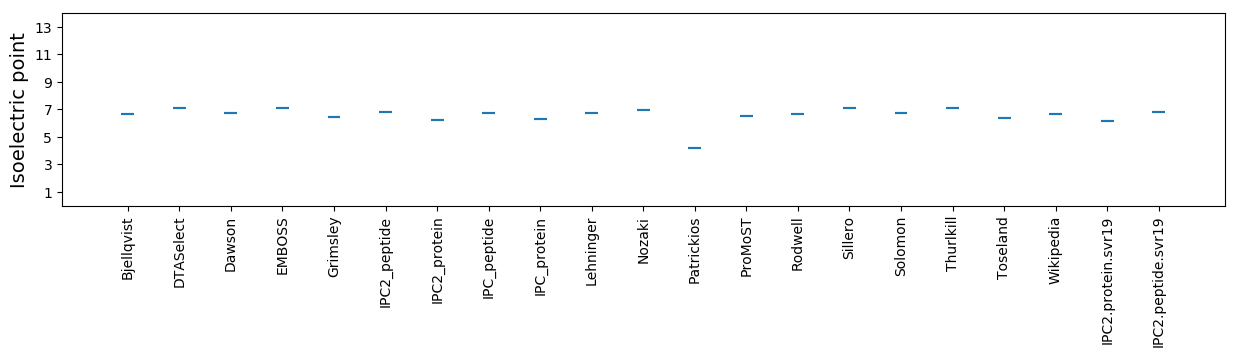
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D3HTY0|D3HTY0_9VIRU Replication protein OS=Croton yellow vein mosaic alphasatellite OX=713089 GN=alpha rep PE=4 SV=1
MM1 pKa = 6.91SHH3 pKa = 5.9SRR5 pKa = 11.84NWCFTIFNYY14 pKa = 9.97VLPLFTTLPEE24 pKa = 3.79WANYY28 pKa = 10.27LVFQEE33 pKa = 4.98EE34 pKa = 4.62EE35 pKa = 4.36CPTTKK40 pKa = 9.85KK41 pKa = 10.48RR42 pKa = 11.84HH43 pKa = 4.56IQGYY47 pKa = 9.82VNLKK51 pKa = 9.19RR52 pKa = 11.84NQRR55 pKa = 11.84FAFLKK60 pKa = 10.48KK61 pKa = 9.96KK62 pKa = 10.7LPDD65 pKa = 3.85GSHH68 pKa = 6.86IEE70 pKa = 4.0PCRR73 pKa = 11.84GSASSNRR80 pKa = 11.84DD81 pKa = 3.15YY82 pKa = 10.47CTKK85 pKa = 10.47DD86 pKa = 2.88ASRR89 pKa = 11.84TGGPWEE95 pKa = 4.17FGVFCEE101 pKa = 4.38TGSNKK106 pKa = 9.76RR107 pKa = 11.84KK108 pKa = 7.94TMEE111 pKa = 4.3RR112 pKa = 11.84FQEE115 pKa = 4.28DD116 pKa = 4.08PEE118 pKa = 4.17EE119 pKa = 4.61LRR121 pKa = 11.84LADD124 pKa = 3.63PKK126 pKa = 10.93LYY128 pKa = 10.25RR129 pKa = 11.84RR130 pKa = 11.84CLATKK135 pKa = 10.51VNTEE139 pKa = 3.71FSGLVLPVPDD149 pKa = 5.19RR150 pKa = 11.84PWQLVAQKK158 pKa = 10.95ALDD161 pKa = 3.56QGPDD165 pKa = 3.18DD166 pKa = 4.05RR167 pKa = 11.84TIIWVYY173 pKa = 10.5GSEE176 pKa = 4.1GNEE179 pKa = 4.89GKK181 pKa = 7.15TTWAKK186 pKa = 10.56IKK188 pKa = 10.3IQEE191 pKa = 3.75GWFYY195 pKa = 11.47SRR197 pKa = 11.84GGKK200 pKa = 9.87GEE202 pKa = 4.13NIKK205 pKa = 10.35YY206 pKa = 10.12QYY208 pKa = 10.88AEE210 pKa = 4.16HH211 pKa = 6.98LGHH214 pKa = 6.82CVFDD218 pKa = 3.66IPRR221 pKa = 11.84QVEE224 pKa = 4.24DD225 pKa = 3.31NLQYY229 pKa = 10.79TVLEE233 pKa = 4.5EE234 pKa = 3.95IKK236 pKa = 10.78DD237 pKa = 3.69RR238 pKa = 11.84LIRR241 pKa = 11.84SSKK244 pKa = 10.56YY245 pKa = 10.02EE246 pKa = 4.51PIDD249 pKa = 3.78FNCSDD254 pKa = 3.65KK255 pKa = 11.19VHH257 pKa = 6.57LVVLSNFLPCLDD269 pKa = 3.29VEE271 pKa = 4.56YY272 pKa = 10.84NNRR275 pKa = 11.84GEE277 pKa = 4.25LVKK280 pKa = 10.8KK281 pKa = 10.14PLLSRR286 pKa = 11.84DD287 pKa = 2.91RR288 pKa = 11.84VFIINIDD295 pKa = 3.63EE296 pKa = 4.59SVCGHH301 pKa = 7.3PDD303 pKa = 3.24DD304 pKa = 5.56LKK306 pKa = 11.53NFDD309 pKa = 4.28VYY311 pKa = 11.54LEE313 pKa = 4.05
MM1 pKa = 6.91SHH3 pKa = 5.9SRR5 pKa = 11.84NWCFTIFNYY14 pKa = 9.97VLPLFTTLPEE24 pKa = 3.79WANYY28 pKa = 10.27LVFQEE33 pKa = 4.98EE34 pKa = 4.62EE35 pKa = 4.36CPTTKK40 pKa = 9.85KK41 pKa = 10.48RR42 pKa = 11.84HH43 pKa = 4.56IQGYY47 pKa = 9.82VNLKK51 pKa = 9.19RR52 pKa = 11.84NQRR55 pKa = 11.84FAFLKK60 pKa = 10.48KK61 pKa = 9.96KK62 pKa = 10.7LPDD65 pKa = 3.85GSHH68 pKa = 6.86IEE70 pKa = 4.0PCRR73 pKa = 11.84GSASSNRR80 pKa = 11.84DD81 pKa = 3.15YY82 pKa = 10.47CTKK85 pKa = 10.47DD86 pKa = 2.88ASRR89 pKa = 11.84TGGPWEE95 pKa = 4.17FGVFCEE101 pKa = 4.38TGSNKK106 pKa = 9.76RR107 pKa = 11.84KK108 pKa = 7.94TMEE111 pKa = 4.3RR112 pKa = 11.84FQEE115 pKa = 4.28DD116 pKa = 4.08PEE118 pKa = 4.17EE119 pKa = 4.61LRR121 pKa = 11.84LADD124 pKa = 3.63PKK126 pKa = 10.93LYY128 pKa = 10.25RR129 pKa = 11.84RR130 pKa = 11.84CLATKK135 pKa = 10.51VNTEE139 pKa = 3.71FSGLVLPVPDD149 pKa = 5.19RR150 pKa = 11.84PWQLVAQKK158 pKa = 10.95ALDD161 pKa = 3.56QGPDD165 pKa = 3.18DD166 pKa = 4.05RR167 pKa = 11.84TIIWVYY173 pKa = 10.5GSEE176 pKa = 4.1GNEE179 pKa = 4.89GKK181 pKa = 7.15TTWAKK186 pKa = 10.56IKK188 pKa = 10.3IQEE191 pKa = 3.75GWFYY195 pKa = 11.47SRR197 pKa = 11.84GGKK200 pKa = 9.87GEE202 pKa = 4.13NIKK205 pKa = 10.35YY206 pKa = 10.12QYY208 pKa = 10.88AEE210 pKa = 4.16HH211 pKa = 6.98LGHH214 pKa = 6.82CVFDD218 pKa = 3.66IPRR221 pKa = 11.84QVEE224 pKa = 4.24DD225 pKa = 3.31NLQYY229 pKa = 10.79TVLEE233 pKa = 4.5EE234 pKa = 3.95IKK236 pKa = 10.78DD237 pKa = 3.69RR238 pKa = 11.84LIRR241 pKa = 11.84SSKK244 pKa = 10.56YY245 pKa = 10.02EE246 pKa = 4.51PIDD249 pKa = 3.78FNCSDD254 pKa = 3.65KK255 pKa = 11.19VHH257 pKa = 6.57LVVLSNFLPCLDD269 pKa = 3.29VEE271 pKa = 4.56YY272 pKa = 10.84NNRR275 pKa = 11.84GEE277 pKa = 4.25LVKK280 pKa = 10.8KK281 pKa = 10.14PLLSRR286 pKa = 11.84DD287 pKa = 2.91RR288 pKa = 11.84VFIINIDD295 pKa = 3.63EE296 pKa = 4.59SVCGHH301 pKa = 7.3PDD303 pKa = 3.24DD304 pKa = 5.56LKK306 pKa = 11.53NFDD309 pKa = 4.28VYY311 pKa = 11.54LEE313 pKa = 4.05
Molecular weight: 36.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
313 |
313 |
313 |
313.0 |
36.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.195 ± 0.0 | 3.195 ± 0.0 |
6.39 ± 0.0 | 7.987 ± 0.0 |
5.112 ± 0.0 | 6.07 ± 0.0 |
2.236 ± 0.0 | 4.792 ± 0.0 |
7.348 ± 0.0 | 8.946 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.639 ± 0.0 | 5.431 ± 0.0 |
5.431 ± 0.0 | 3.514 ± 0.0 |
6.709 ± 0.0 | 5.431 ± 0.0 |
4.792 ± 0.0 | 6.39 ± 0.0 |
2.236 ± 0.0 | 4.153 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
