
Dragonfly-associated circular virus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemyduguivirus; Gemyduguivirus draga1; Dragonfly associated gemyduguivirus 1
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
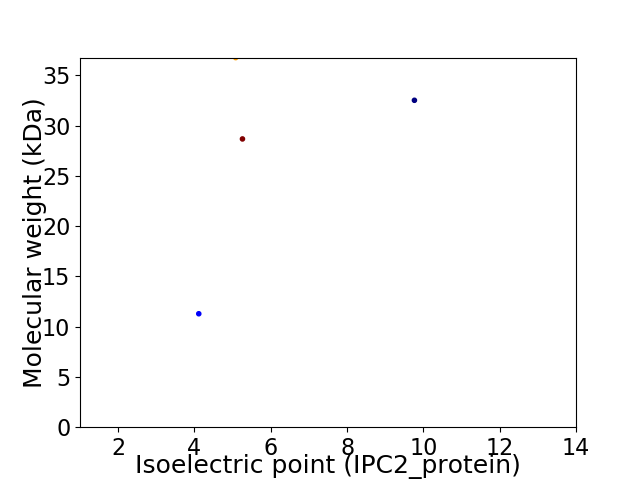
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0A2I7|K0A2I7_9VIRU Replication-associated protein OS=Dragonfly-associated circular virus 3 OX=1234884 PE=3 SV=1
MM1 pKa = 7.82DD2 pKa = 4.3QLSFDD7 pKa = 4.07TDD9 pKa = 3.62DD10 pKa = 3.18VNYY13 pKa = 11.01AVFDD17 pKa = 5.38DD18 pKa = 3.52IHH20 pKa = 7.3SLKK23 pKa = 10.09FFPMWKK29 pKa = 9.42FWMGAQDD36 pKa = 3.75TFTVTDD42 pKa = 3.79KK43 pKa = 11.55YY44 pKa = 10.25KK45 pKa = 11.42GKK47 pKa = 8.14MTFNWGRR54 pKa = 11.84PIIWCNNKK62 pKa = 9.92DD63 pKa = 3.48PRR65 pKa = 11.84ADD67 pKa = 3.71PDD69 pKa = 5.01ADD71 pKa = 4.6ADD73 pKa = 4.32WIDD76 pKa = 3.75ANCIVVNVPEE86 pKa = 4.4DD87 pKa = 3.61TPLISRR93 pKa = 11.84ANTEE97 pKa = 3.79
MM1 pKa = 7.82DD2 pKa = 4.3QLSFDD7 pKa = 4.07TDD9 pKa = 3.62DD10 pKa = 3.18VNYY13 pKa = 11.01AVFDD17 pKa = 5.38DD18 pKa = 3.52IHH20 pKa = 7.3SLKK23 pKa = 10.09FFPMWKK29 pKa = 9.42FWMGAQDD36 pKa = 3.75TFTVTDD42 pKa = 3.79KK43 pKa = 11.55YY44 pKa = 10.25KK45 pKa = 11.42GKK47 pKa = 8.14MTFNWGRR54 pKa = 11.84PIIWCNNKK62 pKa = 9.92DD63 pKa = 3.48PRR65 pKa = 11.84ADD67 pKa = 3.71PDD69 pKa = 5.01ADD71 pKa = 4.6ADD73 pKa = 4.32WIDD76 pKa = 3.75ANCIVVNVPEE86 pKa = 4.4DD87 pKa = 3.61TPLISRR93 pKa = 11.84ANTEE97 pKa = 3.79
Molecular weight: 11.28 kDa
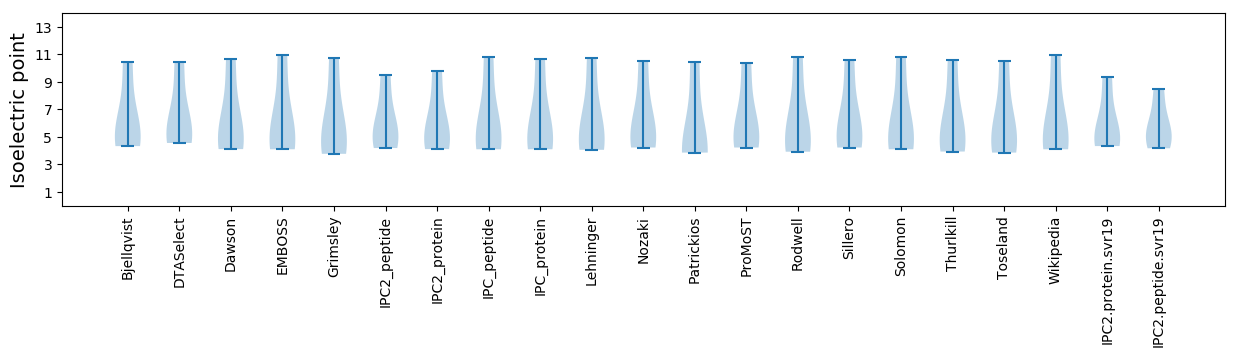
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0A1L3|K0A1L3_9VIRU Replication-associated protein OS=Dragonfly-associated circular virus 3 OX=1234884 PE=4 SV=1
MM1 pKa = 7.82AYY3 pKa = 10.41ASRR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 9.55ARR10 pKa = 11.84RR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84SLRR17 pKa = 11.84TSRR20 pKa = 11.84YY21 pKa = 8.32RR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84GPRR28 pKa = 11.84RR29 pKa = 11.84VRR31 pKa = 11.84RR32 pKa = 11.84TTFRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GTSKK42 pKa = 10.4RR43 pKa = 11.84RR44 pKa = 11.84ILNVSSKK51 pKa = 10.54KK52 pKa = 10.35KK53 pKa = 9.01RR54 pKa = 11.84DD55 pKa = 3.28NMINVSFSPTGINPAVRR72 pKa = 11.84GFIATGAAPTMLVWCATARR91 pKa = 11.84DD92 pKa = 3.89RR93 pKa = 11.84VSNASNPTADD103 pKa = 4.09SVRR106 pKa = 11.84EE107 pKa = 4.02SQTCYY112 pKa = 9.5MRR114 pKa = 11.84GLKK117 pKa = 9.09EE118 pKa = 4.09HH119 pKa = 6.62IRR121 pKa = 11.84INTNNPTSWLWRR133 pKa = 11.84RR134 pKa = 11.84ICFTAKK140 pKa = 10.49GLFSGLGTSADD151 pKa = 3.53SVEE154 pKa = 4.54TSNGWVRR161 pKa = 11.84ALMDD165 pKa = 3.22QTGTSFGTSLTNLLFAGTNGVDD187 pKa = 3.16WNDD190 pKa = 3.12TFTANTDD197 pKa = 3.43STNVKK202 pKa = 9.8IMYY205 pKa = 9.22DD206 pKa = 3.08KK207 pKa = 11.05TFTINSGVSGVIRR220 pKa = 11.84DD221 pKa = 3.55YY222 pKa = 11.89KK223 pKa = 9.47MWHH226 pKa = 5.98GMNKK230 pKa = 9.93NIVYY234 pKa = 10.63GEE236 pKa = 4.25DD237 pKa = 3.4EE238 pKa = 4.27NGEE241 pKa = 4.13GEE243 pKa = 4.09INNRR247 pKa = 11.84YY248 pKa = 7.85STIGRR253 pKa = 11.84PGMGDD258 pKa = 3.69YY259 pKa = 11.13YY260 pKa = 11.47VVDD263 pKa = 4.67LLASNSSAAADD274 pKa = 3.44TLTFSTEE281 pKa = 3.42ATLYY285 pKa = 8.61WHH287 pKa = 7.09EE288 pKa = 4.26KK289 pKa = 9.3
MM1 pKa = 7.82AYY3 pKa = 10.41ASRR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 9.55ARR10 pKa = 11.84RR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84SLRR17 pKa = 11.84TSRR20 pKa = 11.84YY21 pKa = 8.32RR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84GPRR28 pKa = 11.84RR29 pKa = 11.84VRR31 pKa = 11.84RR32 pKa = 11.84TTFRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GTSKK42 pKa = 10.4RR43 pKa = 11.84RR44 pKa = 11.84ILNVSSKK51 pKa = 10.54KK52 pKa = 10.35KK53 pKa = 9.01RR54 pKa = 11.84DD55 pKa = 3.28NMINVSFSPTGINPAVRR72 pKa = 11.84GFIATGAAPTMLVWCATARR91 pKa = 11.84DD92 pKa = 3.89RR93 pKa = 11.84VSNASNPTADD103 pKa = 4.09SVRR106 pKa = 11.84EE107 pKa = 4.02SQTCYY112 pKa = 9.5MRR114 pKa = 11.84GLKK117 pKa = 9.09EE118 pKa = 4.09HH119 pKa = 6.62IRR121 pKa = 11.84INTNNPTSWLWRR133 pKa = 11.84RR134 pKa = 11.84ICFTAKK140 pKa = 10.49GLFSGLGTSADD151 pKa = 3.53SVEE154 pKa = 4.54TSNGWVRR161 pKa = 11.84ALMDD165 pKa = 3.22QTGTSFGTSLTNLLFAGTNGVDD187 pKa = 3.16WNDD190 pKa = 3.12TFTANTDD197 pKa = 3.43STNVKK202 pKa = 9.8IMYY205 pKa = 9.22DD206 pKa = 3.08KK207 pKa = 11.05TFTINSGVSGVIRR220 pKa = 11.84DD221 pKa = 3.55YY222 pKa = 11.89KK223 pKa = 9.47MWHH226 pKa = 5.98GMNKK230 pKa = 9.93NIVYY234 pKa = 10.63GEE236 pKa = 4.25DD237 pKa = 3.4EE238 pKa = 4.27NGEE241 pKa = 4.13GEE243 pKa = 4.09INNRR247 pKa = 11.84YY248 pKa = 7.85STIGRR253 pKa = 11.84PGMGDD258 pKa = 3.69YY259 pKa = 11.13YY260 pKa = 11.47VVDD263 pKa = 4.67LLASNSSAAADD274 pKa = 3.44TLTFSTEE281 pKa = 3.42ATLYY285 pKa = 8.61WHH287 pKa = 7.09EE288 pKa = 4.26KK289 pKa = 9.3
Molecular weight: 32.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
965 |
97 |
323 |
241.3 |
27.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.254 ± 0.206 | 1.554 ± 0.192 |
7.565 ± 1.618 | 4.56 ± 0.778 |
5.699 ± 0.814 | 7.979 ± 0.805 |
1.451 ± 0.199 | 4.456 ± 0.294 |
4.145 ± 0.594 | 5.803 ± 0.567 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.798 ± 0.266 | 5.389 ± 0.994 |
4.974 ± 0.849 | 2.28 ± 0.651 |
7.668 ± 1.446 | 7.15 ± 0.918 |
7.358 ± 1.409 | 5.285 ± 0.227 |
2.798 ± 0.457 | 3.834 ± 0.287 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
