
Euphorbia heterophylla associated gemycircularvirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus euhet1
Average proteome isoelectric point is 7.47
Get precalculated fractions of proteins
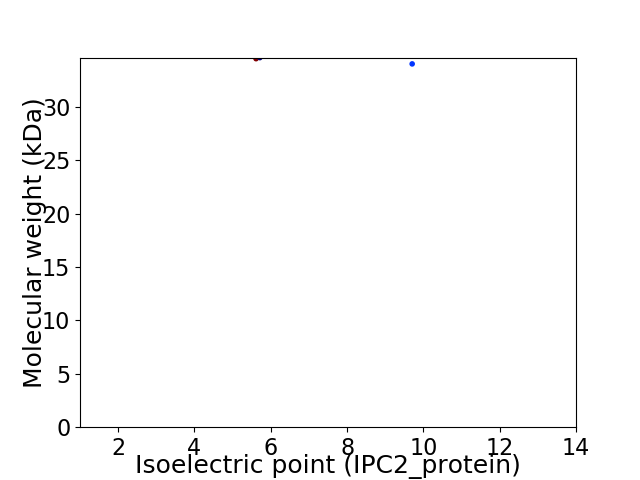
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345S7V1|A0A345S7V1_9VIRU Replication-associated protein OS=Euphorbia heterophylla associated gemycircularvirus OX=2291614 PE=3 SV=1
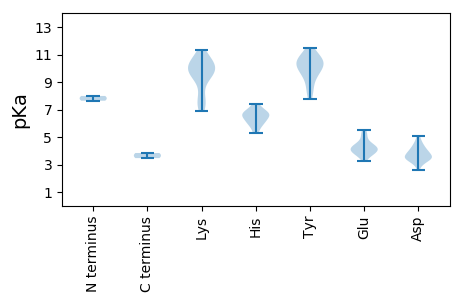
MM1 pKa = 7.64SFTTNGRR8 pKa = 11.84YY9 pKa = 9.13FLVTYY14 pKa = 7.77AQCGDD19 pKa = 4.01LDD21 pKa = 3.89PFRR24 pKa = 11.84VVDD27 pKa = 3.95KK28 pKa = 10.74FSSLGSEE35 pKa = 4.34CIIGRR40 pKa = 11.84EE41 pKa = 3.92LHH43 pKa = 6.64EE44 pKa = 5.48DD45 pKa = 3.55GGLHH49 pKa = 5.93LHH51 pKa = 6.61CFVDD55 pKa = 4.63FGRR58 pKa = 11.84KK59 pKa = 8.02FRR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 7.79TDD66 pKa = 2.98VFDD69 pKa = 3.81VDD71 pKa = 3.68GRR73 pKa = 11.84HH74 pKa = 6.13PNIEE78 pKa = 4.17SSRR81 pKa = 11.84GTPEE85 pKa = 3.23KK86 pKa = 10.53GYY88 pKa = 11.15NYY90 pKa = 9.99AIKK93 pKa = 10.82DD94 pKa = 3.54GDD96 pKa = 4.0VVAGGLQRR104 pKa = 11.84PEE106 pKa = 3.91PKK108 pKa = 10.31SRR110 pKa = 11.84DD111 pKa = 3.45GAGSTFEE118 pKa = 3.98KK119 pKa = 10.22WSLITSATDD128 pKa = 3.42RR129 pKa = 11.84EE130 pKa = 4.54EE131 pKa = 3.71FWRR134 pKa = 11.84LVHH137 pKa = 6.84EE138 pKa = 5.35LDD140 pKa = 4.34PKK142 pKa = 10.76SAACSFTQLSKK153 pKa = 11.32YY154 pKa = 10.58ADD156 pKa = 3.33SKK158 pKa = 9.27YY159 pKa = 11.04AEE161 pKa = 4.23VPTEE165 pKa = 3.99YY166 pKa = 9.9EE167 pKa = 3.92HH168 pKa = 7.4PGGIEE173 pKa = 3.7FAPGDD178 pKa = 4.39GRR180 pKa = 11.84PMSLVLYY187 pKa = 10.34GEE189 pKa = 4.26SRR191 pKa = 11.84TGKK194 pKa = 7.35TLWARR199 pKa = 11.84SLGRR203 pKa = 11.84HH204 pKa = 6.0IYY206 pKa = 10.36NVGLVSGAEE215 pKa = 4.06CVKK218 pKa = 10.88ASEE221 pKa = 3.66VDD223 pKa = 3.53YY224 pKa = 11.45AVFDD228 pKa = 5.08DD229 pKa = 4.23IRR231 pKa = 11.84GGMKK235 pKa = 9.7FFPAFKK241 pKa = 9.52EE242 pKa = 4.07WLGGQHH248 pKa = 5.3TVCVKK253 pKa = 10.12QLYY256 pKa = 9.69RR257 pKa = 11.84DD258 pKa = 4.33PILVKK263 pKa = 9.81WGKK266 pKa = 9.37PSIWVSNDD274 pKa = 3.04DD275 pKa = 4.09PRR277 pKa = 11.84LCMEE281 pKa = 5.06PSDD284 pKa = 4.56VAWLEE289 pKa = 3.74ANARR293 pKa = 11.84FIEE296 pKa = 4.52CNHH299 pKa = 6.77AIFRR303 pKa = 11.84ANTEE307 pKa = 3.84
MM1 pKa = 7.64SFTTNGRR8 pKa = 11.84YY9 pKa = 9.13FLVTYY14 pKa = 7.77AQCGDD19 pKa = 4.01LDD21 pKa = 3.89PFRR24 pKa = 11.84VVDD27 pKa = 3.95KK28 pKa = 10.74FSSLGSEE35 pKa = 4.34CIIGRR40 pKa = 11.84EE41 pKa = 3.92LHH43 pKa = 6.64EE44 pKa = 5.48DD45 pKa = 3.55GGLHH49 pKa = 5.93LHH51 pKa = 6.61CFVDD55 pKa = 4.63FGRR58 pKa = 11.84KK59 pKa = 8.02FRR61 pKa = 11.84SRR63 pKa = 11.84KK64 pKa = 7.79TDD66 pKa = 2.98VFDD69 pKa = 3.81VDD71 pKa = 3.68GRR73 pKa = 11.84HH74 pKa = 6.13PNIEE78 pKa = 4.17SSRR81 pKa = 11.84GTPEE85 pKa = 3.23KK86 pKa = 10.53GYY88 pKa = 11.15NYY90 pKa = 9.99AIKK93 pKa = 10.82DD94 pKa = 3.54GDD96 pKa = 4.0VVAGGLQRR104 pKa = 11.84PEE106 pKa = 3.91PKK108 pKa = 10.31SRR110 pKa = 11.84DD111 pKa = 3.45GAGSTFEE118 pKa = 3.98KK119 pKa = 10.22WSLITSATDD128 pKa = 3.42RR129 pKa = 11.84EE130 pKa = 4.54EE131 pKa = 3.71FWRR134 pKa = 11.84LVHH137 pKa = 6.84EE138 pKa = 5.35LDD140 pKa = 4.34PKK142 pKa = 10.76SAACSFTQLSKK153 pKa = 11.32YY154 pKa = 10.58ADD156 pKa = 3.33SKK158 pKa = 9.27YY159 pKa = 11.04AEE161 pKa = 4.23VPTEE165 pKa = 3.99YY166 pKa = 9.9EE167 pKa = 3.92HH168 pKa = 7.4PGGIEE173 pKa = 3.7FAPGDD178 pKa = 4.39GRR180 pKa = 11.84PMSLVLYY187 pKa = 10.34GEE189 pKa = 4.26SRR191 pKa = 11.84TGKK194 pKa = 7.35TLWARR199 pKa = 11.84SLGRR203 pKa = 11.84HH204 pKa = 6.0IYY206 pKa = 10.36NVGLVSGAEE215 pKa = 4.06CVKK218 pKa = 10.88ASEE221 pKa = 3.66VDD223 pKa = 3.53YY224 pKa = 11.45AVFDD228 pKa = 5.08DD229 pKa = 4.23IRR231 pKa = 11.84GGMKK235 pKa = 9.7FFPAFKK241 pKa = 9.52EE242 pKa = 4.07WLGGQHH248 pKa = 5.3TVCVKK253 pKa = 10.12QLYY256 pKa = 9.69RR257 pKa = 11.84DD258 pKa = 4.33PILVKK263 pKa = 9.81WGKK266 pKa = 9.37PSIWVSNDD274 pKa = 3.04DD275 pKa = 4.09PRR277 pKa = 11.84LCMEE281 pKa = 5.06PSDD284 pKa = 4.56VAWLEE289 pKa = 3.74ANARR293 pKa = 11.84FIEE296 pKa = 4.52CNHH299 pKa = 6.77AIFRR303 pKa = 11.84ANTEE307 pKa = 3.84
Molecular weight: 34.5 kDa
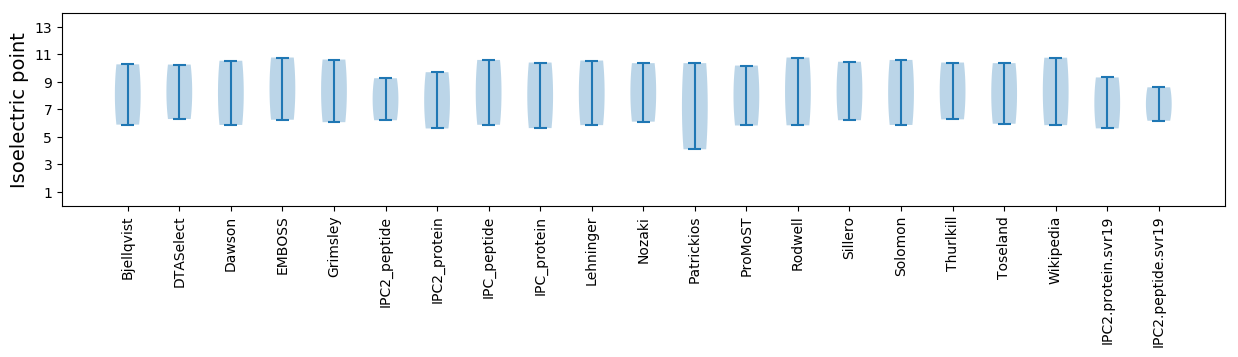
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345S7V1|A0A345S7V1_9VIRU Replication-associated protein OS=Euphorbia heterophylla associated gemycircularvirus OX=2291614 PE=3 SV=1
MM1 pKa = 7.99PYY3 pKa = 9.67RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 10.07ARR8 pKa = 11.84KK9 pKa = 7.4PARR12 pKa = 11.84NRR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.38GGVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 9.07PIKK26 pKa = 9.51KK27 pKa = 6.92TQFRR31 pKa = 11.84KK32 pKa = 10.17RR33 pKa = 11.84RR34 pKa = 11.84MTPRR38 pKa = 11.84KK39 pKa = 9.48RR40 pKa = 11.84ILNIASRR47 pKa = 11.84KK48 pKa = 9.52KK49 pKa = 9.71RR50 pKa = 11.84DD51 pKa = 3.38NMIANTNITISPKK64 pKa = 10.15PGTFASGGAVLTGDD78 pKa = 3.62RR79 pKa = 11.84QYY81 pKa = 10.93IIPWVASGRR90 pKa = 11.84PLNGSDD96 pKa = 3.14NTNNTIAEE104 pKa = 4.2AAARR108 pKa = 11.84TATEE112 pKa = 4.21CYY114 pKa = 9.52MRR116 pKa = 11.84GLNEE120 pKa = 4.62KK121 pKa = 9.97IQIQTSTAQPWQWRR135 pKa = 11.84RR136 pKa = 11.84VCFQYY141 pKa = 10.42RR142 pKa = 11.84QRR144 pKa = 11.84DD145 pKa = 3.46ILNTQNADD153 pKa = 3.59TPLVAQTAAVGLTRR167 pKa = 11.84GLVDD171 pKa = 3.96ALQNGTMAQTLLNVLFQGRR190 pKa = 11.84QSIDD194 pKa = 2.63WASFFTAKK202 pKa = 10.01VDD204 pKa = 3.56NRR206 pKa = 11.84NVTLCYY212 pKa = 10.55DD213 pKa = 3.78RR214 pKa = 11.84TRR216 pKa = 11.84IINSGNNNGIIRR228 pKa = 11.84NYY230 pKa = 9.91KK231 pKa = 8.65IWHH234 pKa = 6.46PMNKK238 pKa = 7.31TLRR241 pKa = 11.84YY242 pKa = 10.16GDD244 pKa = 4.87DD245 pKa = 3.22EE246 pKa = 4.59TGNDD250 pKa = 3.29EE251 pKa = 4.34TSSFFCSASNYY262 pKa = 10.42GMGDD266 pKa = 3.54YY267 pKa = 10.78YY268 pKa = 10.49IVDD271 pKa = 3.91IFSAGPSGTSNDD283 pKa = 3.4QLKK286 pKa = 10.38FEE288 pKa = 5.03PEE290 pKa = 3.5ATLYY294 pKa = 8.42WHH296 pKa = 6.69EE297 pKa = 4.18RR298 pKa = 3.47
MM1 pKa = 7.99PYY3 pKa = 9.67RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 10.07ARR8 pKa = 11.84KK9 pKa = 7.4PARR12 pKa = 11.84NRR14 pKa = 11.84RR15 pKa = 11.84YY16 pKa = 8.38GGVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84YY23 pKa = 9.07PIKK26 pKa = 9.51KK27 pKa = 6.92TQFRR31 pKa = 11.84KK32 pKa = 10.17RR33 pKa = 11.84RR34 pKa = 11.84MTPRR38 pKa = 11.84KK39 pKa = 9.48RR40 pKa = 11.84ILNIASRR47 pKa = 11.84KK48 pKa = 9.52KK49 pKa = 9.71RR50 pKa = 11.84DD51 pKa = 3.38NMIANTNITISPKK64 pKa = 10.15PGTFASGGAVLTGDD78 pKa = 3.62RR79 pKa = 11.84QYY81 pKa = 10.93IIPWVASGRR90 pKa = 11.84PLNGSDD96 pKa = 3.14NTNNTIAEE104 pKa = 4.2AAARR108 pKa = 11.84TATEE112 pKa = 4.21CYY114 pKa = 9.52MRR116 pKa = 11.84GLNEE120 pKa = 4.62KK121 pKa = 9.97IQIQTSTAQPWQWRR135 pKa = 11.84RR136 pKa = 11.84VCFQYY141 pKa = 10.42RR142 pKa = 11.84QRR144 pKa = 11.84DD145 pKa = 3.46ILNTQNADD153 pKa = 3.59TPLVAQTAAVGLTRR167 pKa = 11.84GLVDD171 pKa = 3.96ALQNGTMAQTLLNVLFQGRR190 pKa = 11.84QSIDD194 pKa = 2.63WASFFTAKK202 pKa = 10.01VDD204 pKa = 3.56NRR206 pKa = 11.84NVTLCYY212 pKa = 10.55DD213 pKa = 3.78RR214 pKa = 11.84TRR216 pKa = 11.84IINSGNNNGIIRR228 pKa = 11.84NYY230 pKa = 9.91KK231 pKa = 8.65IWHH234 pKa = 6.46PMNKK238 pKa = 7.31TLRR241 pKa = 11.84YY242 pKa = 10.16GDD244 pKa = 4.87DD245 pKa = 3.22EE246 pKa = 4.59TGNDD250 pKa = 3.29EE251 pKa = 4.34TSSFFCSASNYY262 pKa = 10.42GMGDD266 pKa = 3.54YY267 pKa = 10.78YY268 pKa = 10.49IVDD271 pKa = 3.91IFSAGPSGTSNDD283 pKa = 3.4QLKK286 pKa = 10.38FEE288 pKa = 5.03PEE290 pKa = 3.5ATLYY294 pKa = 8.42WHH296 pKa = 6.69EE297 pKa = 4.18RR298 pKa = 3.47
Molecular weight: 34.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605 |
298 |
307 |
302.5 |
34.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.273 ± 0.527 | 1.983 ± 0.433 |
6.116 ± 0.731 | 4.959 ± 1.536 |
4.628 ± 0.859 | 8.264 ± 0.822 |
1.818 ± 0.775 | 5.289 ± 0.96 |
5.124 ± 0.288 | 6.116 ± 0.504 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.818 ± 0.358 | 5.455 ± 1.982 |
4.628 ± 0.179 | 3.306 ± 1.167 |
8.76 ± 1.336 | 6.446 ± 0.727 |
6.612 ± 1.427 | 5.289 ± 1.079 |
2.149 ± 0.091 | 3.967 ± 0.267 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
