
Chenopodium quinoa mitovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Howeltoviricetes; Cryppavirales; Mitoviridae; Mitovirus; unclassified Mitovirus
Average proteome isoelectric point is 9.18
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U8LM99|A0A2U8LM99_9VIRU RNA dependent RNA polymerase OS=Chenopodium quinoa mitovirus 1 OX=2185087 PE=4 SV=1
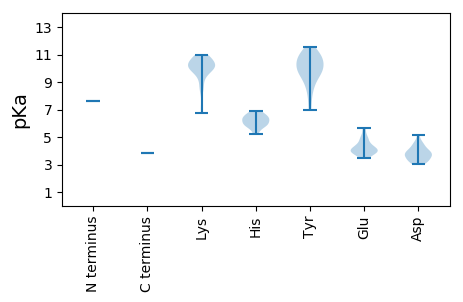
MM1 pKa = 7.61IKK3 pKa = 10.58NLFLNAEE10 pKa = 4.18RR11 pKa = 11.84VPILSWRR18 pKa = 11.84SIFEE22 pKa = 3.61GTRR25 pKa = 11.84RR26 pKa = 11.84LKK28 pKa = 10.63GLCLRR33 pKa = 11.84ASLIVSLRR41 pKa = 11.84LHH43 pKa = 6.66KK44 pKa = 9.83EE45 pKa = 3.5TGLAAVAFARR55 pKa = 11.84VVFRR59 pKa = 11.84LVRR62 pKa = 11.84KK63 pKa = 10.11SGLLFTSLYY72 pKa = 10.53LKK74 pKa = 10.4QCGVSLQRR82 pKa = 11.84YY83 pKa = 6.94YY84 pKa = 11.18AGSYY88 pKa = 10.07SKK90 pKa = 10.52QDD92 pKa = 3.54SLSVPVSLTRR102 pKa = 11.84TGLPKK107 pKa = 10.12IIPVYY112 pKa = 8.98IRR114 pKa = 11.84RR115 pKa = 11.84VIRR118 pKa = 11.84KK119 pKa = 9.13NDD121 pKa = 3.12DD122 pKa = 3.62RR123 pKa = 11.84ADD125 pKa = 3.28KK126 pKa = 10.12WVRR129 pKa = 11.84TYY131 pKa = 11.23LSWFSASRR139 pKa = 11.84LVEE142 pKa = 3.95LAPSVTSSTFASIHH156 pKa = 5.85EE157 pKa = 4.48PIKK160 pKa = 10.9DD161 pKa = 3.04IGSVKK166 pKa = 10.23EE167 pKa = 3.94VLSVLKK173 pKa = 10.14MRR175 pKa = 11.84GRR177 pKa = 11.84RR178 pKa = 11.84LVSTYY183 pKa = 11.05LPFARR188 pKa = 11.84SIPVYY193 pKa = 10.24QGLKK197 pKa = 9.54WEE199 pKa = 4.61PTWKK203 pKa = 8.5STPMTTKK210 pKa = 9.62FVSRR214 pKa = 11.84YY215 pKa = 9.38RR216 pKa = 11.84SLTPEE221 pKa = 4.11EE222 pKa = 5.52LSTLDD227 pKa = 3.52DD228 pKa = 3.8TNIFSNLKK236 pKa = 10.16HH237 pKa = 5.88EE238 pKa = 4.95LASFMYY244 pKa = 10.25NINKK248 pKa = 8.66IHH250 pKa = 6.55SLPEE254 pKa = 4.08GFFSPGCLWPEE265 pKa = 4.02WIIYY269 pKa = 9.51PFDD272 pKa = 3.94FKK274 pKa = 10.83RR275 pKa = 11.84TTEE278 pKa = 3.67IANWSLEE285 pKa = 3.82WFEE288 pKa = 4.98RR289 pKa = 11.84RR290 pKa = 11.84IGPHH294 pKa = 5.25MSSIVSAYY302 pKa = 9.74QIHH305 pKa = 6.86PPVPLFTGKK314 pKa = 10.1LAQTLPGAGKK324 pKa = 10.01RR325 pKa = 11.84RR326 pKa = 11.84IFAICNYY333 pKa = 8.29VKK335 pKa = 10.68QMLLKK340 pKa = 10.04PIHH343 pKa = 5.8SWAMKK348 pKa = 9.19ILSSIPMDD356 pKa = 3.29GTFNQVAPLLRR367 pKa = 11.84LAKK370 pKa = 10.01IKK372 pKa = 9.4RR373 pKa = 11.84THH375 pKa = 6.33VYY377 pKa = 10.79SFDD380 pKa = 3.81LKK382 pKa = 10.88SATDD386 pKa = 3.7RR387 pKa = 11.84WPLPIIYY394 pKa = 8.59TCVASFFGEE403 pKa = 4.89TYY405 pKa = 10.54ASSVVNSTLGLNTFRR420 pKa = 11.84VDD422 pKa = 3.21KK423 pKa = 10.3PIVSRR428 pKa = 11.84MSEE431 pKa = 3.5IAFRR435 pKa = 11.84CGQPLGYY442 pKa = 9.66YY443 pKa = 10.17GSWSLFALSHH453 pKa = 6.41HH454 pKa = 5.92LVVWLAADD462 pKa = 4.01LAYY465 pKa = 9.79PSRR468 pKa = 11.84EE469 pKa = 4.01TPFRR473 pKa = 11.84DD474 pKa = 3.62YY475 pKa = 11.52AVLGDD480 pKa = 4.6DD481 pKa = 5.12VLIADD486 pKa = 4.03TNVALEE492 pKa = 4.12YY493 pKa = 10.65KK494 pKa = 10.42SLLSRR499 pKa = 11.84LGVSISEE506 pKa = 4.31SKK508 pKa = 10.88SIISEE513 pKa = 3.85TGAIEE518 pKa = 3.9FAKK521 pKa = 10.29KK522 pKa = 9.85YY523 pKa = 7.65WVKK526 pKa = 10.85GMQVDD531 pKa = 4.51LSPVSLKK538 pKa = 10.82SLLGARR544 pKa = 11.84HH545 pKa = 6.28TIGLCQIGAKK555 pKa = 9.88YY556 pKa = 10.76DD557 pKa = 3.77LDD559 pKa = 4.3FNTLLRR565 pKa = 11.84IGGAGYY571 pKa = 10.07RR572 pKa = 11.84VRR574 pKa = 11.84SRR576 pKa = 11.84QLSTLNRR583 pKa = 11.84KK584 pKa = 6.75WEE586 pKa = 4.38KK587 pKa = 10.31IRR589 pKa = 11.84AVQVKK594 pKa = 8.3LTGQRR599 pKa = 11.84AQLPIEE605 pKa = 4.08FWIGRR610 pKa = 11.84GCPLDD615 pKa = 4.31PYY617 pKa = 10.68LKK619 pKa = 10.65GKK621 pKa = 10.1IIVYY625 pKa = 9.45LRR627 pKa = 11.84KK628 pKa = 9.56EE629 pKa = 4.35FKK631 pKa = 10.32PKK633 pKa = 9.99EE634 pKa = 3.93LRR636 pKa = 11.84LTPEE640 pKa = 4.0GLLFDD645 pKa = 4.6GEE647 pKa = 4.47VEE649 pKa = 3.88IAEE652 pKa = 4.04RR653 pKa = 11.84TVIHH657 pKa = 6.21NWMKK661 pKa = 10.61QWLKK665 pKa = 9.8WIYY668 pKa = 8.85WYY670 pKa = 11.05YY671 pKa = 10.25SVAMSPDD678 pKa = 3.1VKK680 pKa = 10.72LDD682 pKa = 3.32QFFDD686 pKa = 3.72APICATSWKK695 pKa = 10.12RR696 pKa = 11.84NQIDD700 pKa = 4.07LNLKK704 pKa = 9.98RR705 pKa = 11.84FSALWRR711 pKa = 11.84CYY713 pKa = 11.03DD714 pKa = 4.07MAVGWPKK721 pKa = 10.61NYY723 pKa = 9.68PWVLPATTVLKK734 pKa = 10.13IDD736 pKa = 3.78EE737 pKa = 4.94LIKK740 pKa = 11.0GGFSGTDD747 pKa = 3.42FLMKK751 pKa = 9.96PVEE754 pKa = 5.66LIVQQNKK761 pKa = 7.23GRR763 pKa = 11.84KK764 pKa = 8.64YY765 pKa = 9.81II766 pKa = 3.87
MM1 pKa = 7.61IKK3 pKa = 10.58NLFLNAEE10 pKa = 4.18RR11 pKa = 11.84VPILSWRR18 pKa = 11.84SIFEE22 pKa = 3.61GTRR25 pKa = 11.84RR26 pKa = 11.84LKK28 pKa = 10.63GLCLRR33 pKa = 11.84ASLIVSLRR41 pKa = 11.84LHH43 pKa = 6.66KK44 pKa = 9.83EE45 pKa = 3.5TGLAAVAFARR55 pKa = 11.84VVFRR59 pKa = 11.84LVRR62 pKa = 11.84KK63 pKa = 10.11SGLLFTSLYY72 pKa = 10.53LKK74 pKa = 10.4QCGVSLQRR82 pKa = 11.84YY83 pKa = 6.94YY84 pKa = 11.18AGSYY88 pKa = 10.07SKK90 pKa = 10.52QDD92 pKa = 3.54SLSVPVSLTRR102 pKa = 11.84TGLPKK107 pKa = 10.12IIPVYY112 pKa = 8.98IRR114 pKa = 11.84RR115 pKa = 11.84VIRR118 pKa = 11.84KK119 pKa = 9.13NDD121 pKa = 3.12DD122 pKa = 3.62RR123 pKa = 11.84ADD125 pKa = 3.28KK126 pKa = 10.12WVRR129 pKa = 11.84TYY131 pKa = 11.23LSWFSASRR139 pKa = 11.84LVEE142 pKa = 3.95LAPSVTSSTFASIHH156 pKa = 5.85EE157 pKa = 4.48PIKK160 pKa = 10.9DD161 pKa = 3.04IGSVKK166 pKa = 10.23EE167 pKa = 3.94VLSVLKK173 pKa = 10.14MRR175 pKa = 11.84GRR177 pKa = 11.84RR178 pKa = 11.84LVSTYY183 pKa = 11.05LPFARR188 pKa = 11.84SIPVYY193 pKa = 10.24QGLKK197 pKa = 9.54WEE199 pKa = 4.61PTWKK203 pKa = 8.5STPMTTKK210 pKa = 9.62FVSRR214 pKa = 11.84YY215 pKa = 9.38RR216 pKa = 11.84SLTPEE221 pKa = 4.11EE222 pKa = 5.52LSTLDD227 pKa = 3.52DD228 pKa = 3.8TNIFSNLKK236 pKa = 10.16HH237 pKa = 5.88EE238 pKa = 4.95LASFMYY244 pKa = 10.25NINKK248 pKa = 8.66IHH250 pKa = 6.55SLPEE254 pKa = 4.08GFFSPGCLWPEE265 pKa = 4.02WIIYY269 pKa = 9.51PFDD272 pKa = 3.94FKK274 pKa = 10.83RR275 pKa = 11.84TTEE278 pKa = 3.67IANWSLEE285 pKa = 3.82WFEE288 pKa = 4.98RR289 pKa = 11.84RR290 pKa = 11.84IGPHH294 pKa = 5.25MSSIVSAYY302 pKa = 9.74QIHH305 pKa = 6.86PPVPLFTGKK314 pKa = 10.1LAQTLPGAGKK324 pKa = 10.01RR325 pKa = 11.84RR326 pKa = 11.84IFAICNYY333 pKa = 8.29VKK335 pKa = 10.68QMLLKK340 pKa = 10.04PIHH343 pKa = 5.8SWAMKK348 pKa = 9.19ILSSIPMDD356 pKa = 3.29GTFNQVAPLLRR367 pKa = 11.84LAKK370 pKa = 10.01IKK372 pKa = 9.4RR373 pKa = 11.84THH375 pKa = 6.33VYY377 pKa = 10.79SFDD380 pKa = 3.81LKK382 pKa = 10.88SATDD386 pKa = 3.7RR387 pKa = 11.84WPLPIIYY394 pKa = 8.59TCVASFFGEE403 pKa = 4.89TYY405 pKa = 10.54ASSVVNSTLGLNTFRR420 pKa = 11.84VDD422 pKa = 3.21KK423 pKa = 10.3PIVSRR428 pKa = 11.84MSEE431 pKa = 3.5IAFRR435 pKa = 11.84CGQPLGYY442 pKa = 9.66YY443 pKa = 10.17GSWSLFALSHH453 pKa = 6.41HH454 pKa = 5.92LVVWLAADD462 pKa = 4.01LAYY465 pKa = 9.79PSRR468 pKa = 11.84EE469 pKa = 4.01TPFRR473 pKa = 11.84DD474 pKa = 3.62YY475 pKa = 11.52AVLGDD480 pKa = 4.6DD481 pKa = 5.12VLIADD486 pKa = 4.03TNVALEE492 pKa = 4.12YY493 pKa = 10.65KK494 pKa = 10.42SLLSRR499 pKa = 11.84LGVSISEE506 pKa = 4.31SKK508 pKa = 10.88SIISEE513 pKa = 3.85TGAIEE518 pKa = 3.9FAKK521 pKa = 10.29KK522 pKa = 9.85YY523 pKa = 7.65WVKK526 pKa = 10.85GMQVDD531 pKa = 4.51LSPVSLKK538 pKa = 10.82SLLGARR544 pKa = 11.84HH545 pKa = 6.28TIGLCQIGAKK555 pKa = 9.88YY556 pKa = 10.76DD557 pKa = 3.77LDD559 pKa = 4.3FNTLLRR565 pKa = 11.84IGGAGYY571 pKa = 10.07RR572 pKa = 11.84VRR574 pKa = 11.84SRR576 pKa = 11.84QLSTLNRR583 pKa = 11.84KK584 pKa = 6.75WEE586 pKa = 4.38KK587 pKa = 10.31IRR589 pKa = 11.84AVQVKK594 pKa = 8.3LTGQRR599 pKa = 11.84AQLPIEE605 pKa = 4.08FWIGRR610 pKa = 11.84GCPLDD615 pKa = 4.31PYY617 pKa = 10.68LKK619 pKa = 10.65GKK621 pKa = 10.1IIVYY625 pKa = 9.45LRR627 pKa = 11.84KK628 pKa = 9.56EE629 pKa = 4.35FKK631 pKa = 10.32PKK633 pKa = 9.99EE634 pKa = 3.93LRR636 pKa = 11.84LTPEE640 pKa = 4.0GLLFDD645 pKa = 4.6GEE647 pKa = 4.47VEE649 pKa = 3.88IAEE652 pKa = 4.04RR653 pKa = 11.84TVIHH657 pKa = 6.21NWMKK661 pKa = 10.61QWLKK665 pKa = 9.8WIYY668 pKa = 8.85WYY670 pKa = 11.05YY671 pKa = 10.25SVAMSPDD678 pKa = 3.1VKK680 pKa = 10.72LDD682 pKa = 3.32QFFDD686 pKa = 3.72APICATSWKK695 pKa = 10.12RR696 pKa = 11.84NQIDD700 pKa = 4.07LNLKK704 pKa = 9.98RR705 pKa = 11.84FSALWRR711 pKa = 11.84CYY713 pKa = 11.03DD714 pKa = 4.07MAVGWPKK721 pKa = 10.61NYY723 pKa = 9.68PWVLPATTVLKK734 pKa = 10.13IDD736 pKa = 3.78EE737 pKa = 4.94LIKK740 pKa = 11.0GGFSGTDD747 pKa = 3.42FLMKK751 pKa = 9.96PVEE754 pKa = 5.66LIVQQNKK761 pKa = 7.23GRR763 pKa = 11.84KK764 pKa = 8.64YY765 pKa = 9.81II766 pKa = 3.87
Molecular weight: 87.71 kDa
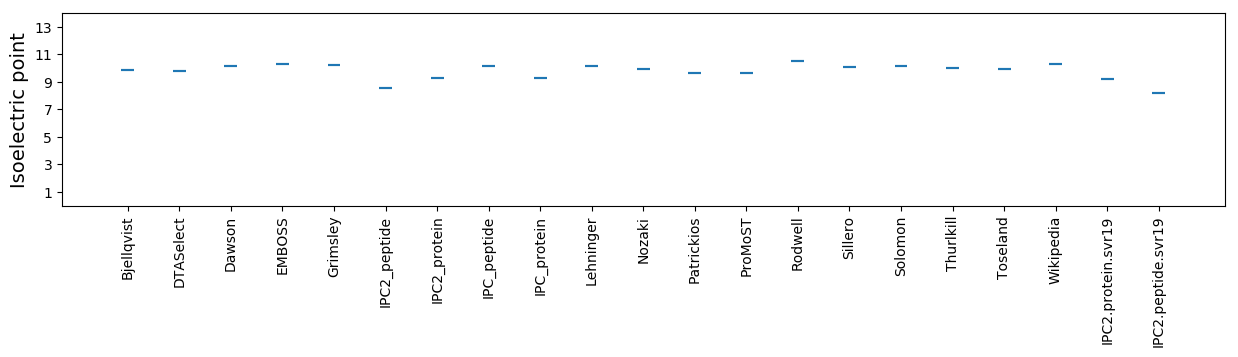
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U8LM99|A0A2U8LM99_9VIRU RNA dependent RNA polymerase OS=Chenopodium quinoa mitovirus 1 OX=2185087 PE=4 SV=1
MM1 pKa = 7.61IKK3 pKa = 10.58NLFLNAEE10 pKa = 4.18RR11 pKa = 11.84VPILSWRR18 pKa = 11.84SIFEE22 pKa = 3.61GTRR25 pKa = 11.84RR26 pKa = 11.84LKK28 pKa = 10.63GLCLRR33 pKa = 11.84ASLIVSLRR41 pKa = 11.84LHH43 pKa = 6.66KK44 pKa = 9.83EE45 pKa = 3.5TGLAAVAFARR55 pKa = 11.84VVFRR59 pKa = 11.84LVRR62 pKa = 11.84KK63 pKa = 10.11SGLLFTSLYY72 pKa = 10.53LKK74 pKa = 10.4QCGVSLQRR82 pKa = 11.84YY83 pKa = 6.94YY84 pKa = 11.18AGSYY88 pKa = 10.07SKK90 pKa = 10.52QDD92 pKa = 3.54SLSVPVSLTRR102 pKa = 11.84TGLPKK107 pKa = 10.12IIPVYY112 pKa = 8.98IRR114 pKa = 11.84RR115 pKa = 11.84VIRR118 pKa = 11.84KK119 pKa = 9.13NDD121 pKa = 3.12DD122 pKa = 3.62RR123 pKa = 11.84ADD125 pKa = 3.28KK126 pKa = 10.12WVRR129 pKa = 11.84TYY131 pKa = 11.23LSWFSASRR139 pKa = 11.84LVEE142 pKa = 3.95LAPSVTSSTFASIHH156 pKa = 5.85EE157 pKa = 4.48PIKK160 pKa = 10.9DD161 pKa = 3.04IGSVKK166 pKa = 10.23EE167 pKa = 3.94VLSVLKK173 pKa = 10.14MRR175 pKa = 11.84GRR177 pKa = 11.84RR178 pKa = 11.84LVSTYY183 pKa = 11.05LPFARR188 pKa = 11.84SIPVYY193 pKa = 10.24QGLKK197 pKa = 9.54WEE199 pKa = 4.61PTWKK203 pKa = 8.5STPMTTKK210 pKa = 9.62FVSRR214 pKa = 11.84YY215 pKa = 9.38RR216 pKa = 11.84SLTPEE221 pKa = 4.11EE222 pKa = 5.52LSTLDD227 pKa = 3.52DD228 pKa = 3.8TNIFSNLKK236 pKa = 10.16HH237 pKa = 5.88EE238 pKa = 4.95LASFMYY244 pKa = 10.25NINKK248 pKa = 8.66IHH250 pKa = 6.55SLPEE254 pKa = 4.08GFFSPGCLWPEE265 pKa = 4.02WIIYY269 pKa = 9.51PFDD272 pKa = 3.94FKK274 pKa = 10.83RR275 pKa = 11.84TTEE278 pKa = 3.67IANWSLEE285 pKa = 3.82WFEE288 pKa = 4.98RR289 pKa = 11.84RR290 pKa = 11.84IGPHH294 pKa = 5.25MSSIVSAYY302 pKa = 9.74QIHH305 pKa = 6.86PPVPLFTGKK314 pKa = 10.1LAQTLPGAGKK324 pKa = 10.01RR325 pKa = 11.84RR326 pKa = 11.84IFAICNYY333 pKa = 8.29VKK335 pKa = 10.68QMLLKK340 pKa = 10.04PIHH343 pKa = 5.8SWAMKK348 pKa = 9.19ILSSIPMDD356 pKa = 3.29GTFNQVAPLLRR367 pKa = 11.84LAKK370 pKa = 10.01IKK372 pKa = 9.4RR373 pKa = 11.84THH375 pKa = 6.33VYY377 pKa = 10.79SFDD380 pKa = 3.81LKK382 pKa = 10.88SATDD386 pKa = 3.7RR387 pKa = 11.84WPLPIIYY394 pKa = 8.59TCVASFFGEE403 pKa = 4.89TYY405 pKa = 10.54ASSVVNSTLGLNTFRR420 pKa = 11.84VDD422 pKa = 3.21KK423 pKa = 10.3PIVSRR428 pKa = 11.84MSEE431 pKa = 3.5IAFRR435 pKa = 11.84CGQPLGYY442 pKa = 9.66YY443 pKa = 10.17GSWSLFALSHH453 pKa = 6.41HH454 pKa = 5.92LVVWLAADD462 pKa = 4.01LAYY465 pKa = 9.79PSRR468 pKa = 11.84EE469 pKa = 4.01TPFRR473 pKa = 11.84DD474 pKa = 3.62YY475 pKa = 11.52AVLGDD480 pKa = 4.6DD481 pKa = 5.12VLIADD486 pKa = 4.03TNVALEE492 pKa = 4.12YY493 pKa = 10.65KK494 pKa = 10.42SLLSRR499 pKa = 11.84LGVSISEE506 pKa = 4.31SKK508 pKa = 10.88SIISEE513 pKa = 3.85TGAIEE518 pKa = 3.9FAKK521 pKa = 10.29KK522 pKa = 9.85YY523 pKa = 7.65WVKK526 pKa = 10.85GMQVDD531 pKa = 4.51LSPVSLKK538 pKa = 10.82SLLGARR544 pKa = 11.84HH545 pKa = 6.28TIGLCQIGAKK555 pKa = 9.88YY556 pKa = 10.76DD557 pKa = 3.77LDD559 pKa = 4.3FNTLLRR565 pKa = 11.84IGGAGYY571 pKa = 10.07RR572 pKa = 11.84VRR574 pKa = 11.84SRR576 pKa = 11.84QLSTLNRR583 pKa = 11.84KK584 pKa = 6.75WEE586 pKa = 4.38KK587 pKa = 10.31IRR589 pKa = 11.84AVQVKK594 pKa = 8.3LTGQRR599 pKa = 11.84AQLPIEE605 pKa = 4.08FWIGRR610 pKa = 11.84GCPLDD615 pKa = 4.31PYY617 pKa = 10.68LKK619 pKa = 10.65GKK621 pKa = 10.1IIVYY625 pKa = 9.45LRR627 pKa = 11.84KK628 pKa = 9.56EE629 pKa = 4.35FKK631 pKa = 10.32PKK633 pKa = 9.99EE634 pKa = 3.93LRR636 pKa = 11.84LTPEE640 pKa = 4.0GLLFDD645 pKa = 4.6GEE647 pKa = 4.47VEE649 pKa = 3.88IAEE652 pKa = 4.04RR653 pKa = 11.84TVIHH657 pKa = 6.21NWMKK661 pKa = 10.61QWLKK665 pKa = 9.8WIYY668 pKa = 8.85WYY670 pKa = 11.05YY671 pKa = 10.25SVAMSPDD678 pKa = 3.1VKK680 pKa = 10.72LDD682 pKa = 3.32QFFDD686 pKa = 3.72APICATSWKK695 pKa = 10.12RR696 pKa = 11.84NQIDD700 pKa = 4.07LNLKK704 pKa = 9.98RR705 pKa = 11.84FSALWRR711 pKa = 11.84CYY713 pKa = 11.03DD714 pKa = 4.07MAVGWPKK721 pKa = 10.61NYY723 pKa = 9.68PWVLPATTVLKK734 pKa = 10.13IDD736 pKa = 3.78EE737 pKa = 4.94LIKK740 pKa = 11.0GGFSGTDD747 pKa = 3.42FLMKK751 pKa = 9.96PVEE754 pKa = 5.66LIVQQNKK761 pKa = 7.23GRR763 pKa = 11.84KK764 pKa = 8.64YY765 pKa = 9.81II766 pKa = 3.87
MM1 pKa = 7.61IKK3 pKa = 10.58NLFLNAEE10 pKa = 4.18RR11 pKa = 11.84VPILSWRR18 pKa = 11.84SIFEE22 pKa = 3.61GTRR25 pKa = 11.84RR26 pKa = 11.84LKK28 pKa = 10.63GLCLRR33 pKa = 11.84ASLIVSLRR41 pKa = 11.84LHH43 pKa = 6.66KK44 pKa = 9.83EE45 pKa = 3.5TGLAAVAFARR55 pKa = 11.84VVFRR59 pKa = 11.84LVRR62 pKa = 11.84KK63 pKa = 10.11SGLLFTSLYY72 pKa = 10.53LKK74 pKa = 10.4QCGVSLQRR82 pKa = 11.84YY83 pKa = 6.94YY84 pKa = 11.18AGSYY88 pKa = 10.07SKK90 pKa = 10.52QDD92 pKa = 3.54SLSVPVSLTRR102 pKa = 11.84TGLPKK107 pKa = 10.12IIPVYY112 pKa = 8.98IRR114 pKa = 11.84RR115 pKa = 11.84VIRR118 pKa = 11.84KK119 pKa = 9.13NDD121 pKa = 3.12DD122 pKa = 3.62RR123 pKa = 11.84ADD125 pKa = 3.28KK126 pKa = 10.12WVRR129 pKa = 11.84TYY131 pKa = 11.23LSWFSASRR139 pKa = 11.84LVEE142 pKa = 3.95LAPSVTSSTFASIHH156 pKa = 5.85EE157 pKa = 4.48PIKK160 pKa = 10.9DD161 pKa = 3.04IGSVKK166 pKa = 10.23EE167 pKa = 3.94VLSVLKK173 pKa = 10.14MRR175 pKa = 11.84GRR177 pKa = 11.84RR178 pKa = 11.84LVSTYY183 pKa = 11.05LPFARR188 pKa = 11.84SIPVYY193 pKa = 10.24QGLKK197 pKa = 9.54WEE199 pKa = 4.61PTWKK203 pKa = 8.5STPMTTKK210 pKa = 9.62FVSRR214 pKa = 11.84YY215 pKa = 9.38RR216 pKa = 11.84SLTPEE221 pKa = 4.11EE222 pKa = 5.52LSTLDD227 pKa = 3.52DD228 pKa = 3.8TNIFSNLKK236 pKa = 10.16HH237 pKa = 5.88EE238 pKa = 4.95LASFMYY244 pKa = 10.25NINKK248 pKa = 8.66IHH250 pKa = 6.55SLPEE254 pKa = 4.08GFFSPGCLWPEE265 pKa = 4.02WIIYY269 pKa = 9.51PFDD272 pKa = 3.94FKK274 pKa = 10.83RR275 pKa = 11.84TTEE278 pKa = 3.67IANWSLEE285 pKa = 3.82WFEE288 pKa = 4.98RR289 pKa = 11.84RR290 pKa = 11.84IGPHH294 pKa = 5.25MSSIVSAYY302 pKa = 9.74QIHH305 pKa = 6.86PPVPLFTGKK314 pKa = 10.1LAQTLPGAGKK324 pKa = 10.01RR325 pKa = 11.84RR326 pKa = 11.84IFAICNYY333 pKa = 8.29VKK335 pKa = 10.68QMLLKK340 pKa = 10.04PIHH343 pKa = 5.8SWAMKK348 pKa = 9.19ILSSIPMDD356 pKa = 3.29GTFNQVAPLLRR367 pKa = 11.84LAKK370 pKa = 10.01IKK372 pKa = 9.4RR373 pKa = 11.84THH375 pKa = 6.33VYY377 pKa = 10.79SFDD380 pKa = 3.81LKK382 pKa = 10.88SATDD386 pKa = 3.7RR387 pKa = 11.84WPLPIIYY394 pKa = 8.59TCVASFFGEE403 pKa = 4.89TYY405 pKa = 10.54ASSVVNSTLGLNTFRR420 pKa = 11.84VDD422 pKa = 3.21KK423 pKa = 10.3PIVSRR428 pKa = 11.84MSEE431 pKa = 3.5IAFRR435 pKa = 11.84CGQPLGYY442 pKa = 9.66YY443 pKa = 10.17GSWSLFALSHH453 pKa = 6.41HH454 pKa = 5.92LVVWLAADD462 pKa = 4.01LAYY465 pKa = 9.79PSRR468 pKa = 11.84EE469 pKa = 4.01TPFRR473 pKa = 11.84DD474 pKa = 3.62YY475 pKa = 11.52AVLGDD480 pKa = 4.6DD481 pKa = 5.12VLIADD486 pKa = 4.03TNVALEE492 pKa = 4.12YY493 pKa = 10.65KK494 pKa = 10.42SLLSRR499 pKa = 11.84LGVSISEE506 pKa = 4.31SKK508 pKa = 10.88SIISEE513 pKa = 3.85TGAIEE518 pKa = 3.9FAKK521 pKa = 10.29KK522 pKa = 9.85YY523 pKa = 7.65WVKK526 pKa = 10.85GMQVDD531 pKa = 4.51LSPVSLKK538 pKa = 10.82SLLGARR544 pKa = 11.84HH545 pKa = 6.28TIGLCQIGAKK555 pKa = 9.88YY556 pKa = 10.76DD557 pKa = 3.77LDD559 pKa = 4.3FNTLLRR565 pKa = 11.84IGGAGYY571 pKa = 10.07RR572 pKa = 11.84VRR574 pKa = 11.84SRR576 pKa = 11.84QLSTLNRR583 pKa = 11.84KK584 pKa = 6.75WEE586 pKa = 4.38KK587 pKa = 10.31IRR589 pKa = 11.84AVQVKK594 pKa = 8.3LTGQRR599 pKa = 11.84AQLPIEE605 pKa = 4.08FWIGRR610 pKa = 11.84GCPLDD615 pKa = 4.31PYY617 pKa = 10.68LKK619 pKa = 10.65GKK621 pKa = 10.1IIVYY625 pKa = 9.45LRR627 pKa = 11.84KK628 pKa = 9.56EE629 pKa = 4.35FKK631 pKa = 10.32PKK633 pKa = 9.99EE634 pKa = 3.93LRR636 pKa = 11.84LTPEE640 pKa = 4.0GLLFDD645 pKa = 4.6GEE647 pKa = 4.47VEE649 pKa = 3.88IAEE652 pKa = 4.04RR653 pKa = 11.84TVIHH657 pKa = 6.21NWMKK661 pKa = 10.61QWLKK665 pKa = 9.8WIYY668 pKa = 8.85WYY670 pKa = 11.05YY671 pKa = 10.25SVAMSPDD678 pKa = 3.1VKK680 pKa = 10.72LDD682 pKa = 3.32QFFDD686 pKa = 3.72APICATSWKK695 pKa = 10.12RR696 pKa = 11.84NQIDD700 pKa = 4.07LNLKK704 pKa = 9.98RR705 pKa = 11.84FSALWRR711 pKa = 11.84CYY713 pKa = 11.03DD714 pKa = 4.07MAVGWPKK721 pKa = 10.61NYY723 pKa = 9.68PWVLPATTVLKK734 pKa = 10.13IDD736 pKa = 3.78EE737 pKa = 4.94LIKK740 pKa = 11.0GGFSGTDD747 pKa = 3.42FLMKK751 pKa = 9.96PVEE754 pKa = 5.66LIVQQNKK761 pKa = 7.23GRR763 pKa = 11.84KK764 pKa = 8.64YY765 pKa = 9.81II766 pKa = 3.87
Molecular weight: 87.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
766 |
766 |
766 |
766.0 |
87.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.005 ± 0.0 | 1.305 ± 0.0 |
3.786 ± 0.0 | 4.178 ± 0.0 |
4.7 ± 0.0 | 5.614 ± 0.0 |
1.567 ± 0.0 | 6.919 ± 0.0 |
6.919 ± 0.0 | 11.619 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.828 ± 0.0 | 2.611 ± 0.0 |
5.352 ± 0.0 | 2.611 ± 0.0 |
6.919 ± 0.0 | 8.747 ± 0.0 |
5.222 ± 0.0 | 6.789 ± 0.0 |
3.133 ± 0.0 | 4.178 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
