
Tortoise microvirus 85
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
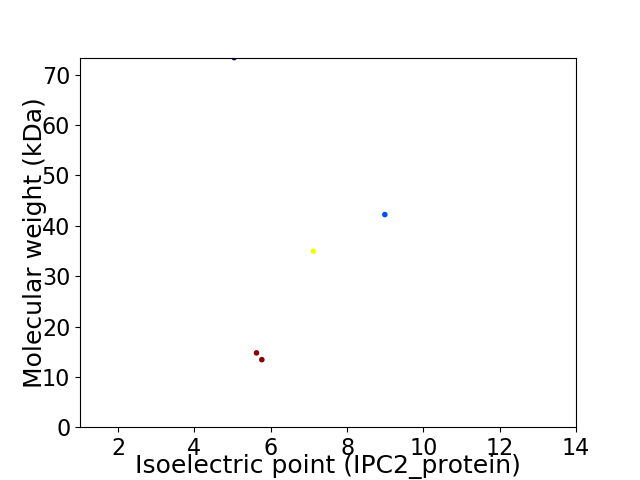
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A4P8W6P6|A0A4P8W6P6_9VIRU DNA pilot protein OS=Tortoise microvirus 85 OX=2583193 PE=4 SV=1
MM1 pKa = 7.57IKK3 pKa = 10.58NLGGDD8 pKa = 3.58RR9 pKa = 11.84LGSGAKK15 pKa = 9.04MNQEE19 pKa = 3.57LHH21 pKa = 5.09GWSRR25 pKa = 11.84STHH28 pKa = 6.88DD29 pKa = 4.12LTTDD33 pKa = 2.85IATTMSIGTLLPIHH47 pKa = 6.73HH48 pKa = 6.48SVILAGDD55 pKa = 3.54TRR57 pKa = 11.84EE58 pKa = 4.0IDD60 pKa = 3.76LDD62 pKa = 3.98AMLLTHH68 pKa = 6.25PTEE71 pKa = 4.9GPLFGSFKK79 pKa = 10.36LQVDD83 pKa = 3.61VFKK86 pKa = 11.36ADD88 pKa = 3.17MRR90 pKa = 11.84LYY92 pKa = 9.97IAKK95 pKa = 10.11LHH97 pKa = 6.05MNLLEE102 pKa = 4.96KK103 pKa = 10.46GLTMSDD109 pKa = 2.51IKK111 pKa = 11.16LPLIEE116 pKa = 4.58MDD118 pKa = 4.44ANNIDD123 pKa = 3.55WTKK126 pKa = 11.34DD127 pKa = 3.23PNNQQINPSCIFKK140 pKa = 10.32YY141 pKa = 10.29LHH143 pKa = 6.61ISGLGYY149 pKa = 10.36NDD151 pKa = 3.46SFEE154 pKa = 4.14APRR157 pKa = 11.84RR158 pKa = 11.84YY159 pKa = 9.98FNAIPWLMYY168 pKa = 8.94WDD170 pKa = 3.74IYY172 pKa = 9.97KK173 pKa = 10.62NYY175 pKa = 9.48YY176 pKa = 9.98ANKK179 pKa = 8.39QEE181 pKa = 4.16TYY183 pKa = 9.95GWVIHH188 pKa = 5.88TGIGLNEE195 pKa = 4.56TITAIKK201 pKa = 9.99IKK203 pKa = 9.03TGSGAEE209 pKa = 4.08STIVEE214 pKa = 4.29APATGLYY221 pKa = 9.95PVTGFTKK228 pKa = 9.5ITIEE232 pKa = 3.95YY233 pKa = 10.54DD234 pKa = 3.24GAQPLLSTVMVNTTNGQQQLDD255 pKa = 3.6QLFTQITYY263 pKa = 10.78EE264 pKa = 4.03NDD266 pKa = 3.15DD267 pKa = 4.47KK268 pKa = 11.36IILEE272 pKa = 4.16IPNSNFNITNWLYY285 pKa = 11.17VEE287 pKa = 4.33NGVPITTVPRR297 pKa = 11.84LEE299 pKa = 4.56KK300 pKa = 10.76FPLTNLDD307 pKa = 4.29EE308 pKa = 5.08IKK310 pKa = 10.06MDD312 pKa = 3.37ILADD316 pKa = 3.54IKK318 pKa = 10.36STSAFTIKK326 pKa = 10.49YY327 pKa = 8.95DD328 pKa = 3.44DD329 pKa = 5.19HH330 pKa = 6.8IAPFSYY336 pKa = 10.74ALEE339 pKa = 4.25LTNGIYY345 pKa = 10.74SKK347 pKa = 10.15MYY349 pKa = 9.17SQEE352 pKa = 3.33GLAIKK357 pKa = 9.28TYY359 pKa = 10.74QSDD362 pKa = 5.1LFNNWLNTEE371 pKa = 4.36YY372 pKa = 10.38IDD374 pKa = 4.63GVNGINARR382 pKa = 11.84TAIQVDD388 pKa = 3.93VNGKK392 pKa = 7.7FTIDD396 pKa = 3.24NFLLKK401 pKa = 10.52EE402 pKa = 3.94KK403 pKa = 10.61LYY405 pKa = 11.26DD406 pKa = 3.48HH407 pKa = 7.37LNRR410 pKa = 11.84IAIAGGTVDD419 pKa = 6.07DD420 pKa = 4.39MMEE423 pKa = 3.84VTYY426 pKa = 10.64DD427 pKa = 3.14VSNRR431 pKa = 11.84TRR433 pKa = 11.84SEE435 pKa = 3.56IPTFEE440 pKa = 5.47GGMSKK445 pKa = 10.3EE446 pKa = 4.01LVFEE450 pKa = 4.03QVISNTATSTQPLGTIAGRR469 pKa = 11.84GTTQGHH475 pKa = 6.27KK476 pKa = 10.04GGKK479 pKa = 8.95IRR481 pKa = 11.84ITNDD485 pKa = 3.13DD486 pKa = 3.82TQHH489 pKa = 6.39AYY491 pKa = 11.31YY492 pKa = 9.65MVIASLTPRR501 pKa = 11.84IKK503 pKa = 10.59YY504 pKa = 9.5SQGNGWDD511 pKa = 4.11TNIKK515 pKa = 9.81TYY517 pKa = 11.33DD518 pKa = 4.12DD519 pKa = 3.52LHH521 pKa = 7.86KK522 pKa = 10.41PAFDD526 pKa = 3.52QIGFQDD532 pKa = 5.58LITDD536 pKa = 4.21QMAFWDD542 pKa = 5.04TYY544 pKa = 10.39CVSGGIAPPVFKK556 pKa = 10.84SAGKK560 pKa = 8.98QPAWINYY567 pKa = 4.47QTEE570 pKa = 3.77IDD572 pKa = 3.52RR573 pKa = 11.84VFGNFAIQSAEE584 pKa = 3.08GWMVLDD590 pKa = 5.9RR591 pKa = 11.84NYY593 pKa = 11.57DD594 pKa = 3.13WDD596 pKa = 4.36AEE598 pKa = 4.13NHH600 pKa = 5.66TIKK603 pKa = 11.01DD604 pKa = 3.6LTTYY608 pKa = 10.51IDD610 pKa = 3.49PEE612 pKa = 4.2KK613 pKa = 10.66YY614 pKa = 10.31NGVFAYY620 pKa = 10.05RR621 pKa = 11.84ALDD624 pKa = 3.67AQNIWAQFKK633 pKa = 10.72VRR635 pKa = 11.84DD636 pKa = 3.47KK637 pKa = 11.15AIRR640 pKa = 11.84KK641 pKa = 8.17MSANQIPRR649 pKa = 11.84MM650 pKa = 4.0
MM1 pKa = 7.57IKK3 pKa = 10.58NLGGDD8 pKa = 3.58RR9 pKa = 11.84LGSGAKK15 pKa = 9.04MNQEE19 pKa = 3.57LHH21 pKa = 5.09GWSRR25 pKa = 11.84STHH28 pKa = 6.88DD29 pKa = 4.12LTTDD33 pKa = 2.85IATTMSIGTLLPIHH47 pKa = 6.73HH48 pKa = 6.48SVILAGDD55 pKa = 3.54TRR57 pKa = 11.84EE58 pKa = 4.0IDD60 pKa = 3.76LDD62 pKa = 3.98AMLLTHH68 pKa = 6.25PTEE71 pKa = 4.9GPLFGSFKK79 pKa = 10.36LQVDD83 pKa = 3.61VFKK86 pKa = 11.36ADD88 pKa = 3.17MRR90 pKa = 11.84LYY92 pKa = 9.97IAKK95 pKa = 10.11LHH97 pKa = 6.05MNLLEE102 pKa = 4.96KK103 pKa = 10.46GLTMSDD109 pKa = 2.51IKK111 pKa = 11.16LPLIEE116 pKa = 4.58MDD118 pKa = 4.44ANNIDD123 pKa = 3.55WTKK126 pKa = 11.34DD127 pKa = 3.23PNNQQINPSCIFKK140 pKa = 10.32YY141 pKa = 10.29LHH143 pKa = 6.61ISGLGYY149 pKa = 10.36NDD151 pKa = 3.46SFEE154 pKa = 4.14APRR157 pKa = 11.84RR158 pKa = 11.84YY159 pKa = 9.98FNAIPWLMYY168 pKa = 8.94WDD170 pKa = 3.74IYY172 pKa = 9.97KK173 pKa = 10.62NYY175 pKa = 9.48YY176 pKa = 9.98ANKK179 pKa = 8.39QEE181 pKa = 4.16TYY183 pKa = 9.95GWVIHH188 pKa = 5.88TGIGLNEE195 pKa = 4.56TITAIKK201 pKa = 9.99IKK203 pKa = 9.03TGSGAEE209 pKa = 4.08STIVEE214 pKa = 4.29APATGLYY221 pKa = 9.95PVTGFTKK228 pKa = 9.5ITIEE232 pKa = 3.95YY233 pKa = 10.54DD234 pKa = 3.24GAQPLLSTVMVNTTNGQQQLDD255 pKa = 3.6QLFTQITYY263 pKa = 10.78EE264 pKa = 4.03NDD266 pKa = 3.15DD267 pKa = 4.47KK268 pKa = 11.36IILEE272 pKa = 4.16IPNSNFNITNWLYY285 pKa = 11.17VEE287 pKa = 4.33NGVPITTVPRR297 pKa = 11.84LEE299 pKa = 4.56KK300 pKa = 10.76FPLTNLDD307 pKa = 4.29EE308 pKa = 5.08IKK310 pKa = 10.06MDD312 pKa = 3.37ILADD316 pKa = 3.54IKK318 pKa = 10.36STSAFTIKK326 pKa = 10.49YY327 pKa = 8.95DD328 pKa = 3.44DD329 pKa = 5.19HH330 pKa = 6.8IAPFSYY336 pKa = 10.74ALEE339 pKa = 4.25LTNGIYY345 pKa = 10.74SKK347 pKa = 10.15MYY349 pKa = 9.17SQEE352 pKa = 3.33GLAIKK357 pKa = 9.28TYY359 pKa = 10.74QSDD362 pKa = 5.1LFNNWLNTEE371 pKa = 4.36YY372 pKa = 10.38IDD374 pKa = 4.63GVNGINARR382 pKa = 11.84TAIQVDD388 pKa = 3.93VNGKK392 pKa = 7.7FTIDD396 pKa = 3.24NFLLKK401 pKa = 10.52EE402 pKa = 3.94KK403 pKa = 10.61LYY405 pKa = 11.26DD406 pKa = 3.48HH407 pKa = 7.37LNRR410 pKa = 11.84IAIAGGTVDD419 pKa = 6.07DD420 pKa = 4.39MMEE423 pKa = 3.84VTYY426 pKa = 10.64DD427 pKa = 3.14VSNRR431 pKa = 11.84TRR433 pKa = 11.84SEE435 pKa = 3.56IPTFEE440 pKa = 5.47GGMSKK445 pKa = 10.3EE446 pKa = 4.01LVFEE450 pKa = 4.03QVISNTATSTQPLGTIAGRR469 pKa = 11.84GTTQGHH475 pKa = 6.27KK476 pKa = 10.04GGKK479 pKa = 8.95IRR481 pKa = 11.84ITNDD485 pKa = 3.13DD486 pKa = 3.82TQHH489 pKa = 6.39AYY491 pKa = 11.31YY492 pKa = 9.65MVIASLTPRR501 pKa = 11.84IKK503 pKa = 10.59YY504 pKa = 9.5SQGNGWDD511 pKa = 4.11TNIKK515 pKa = 9.81TYY517 pKa = 11.33DD518 pKa = 4.12DD519 pKa = 3.52LHH521 pKa = 7.86KK522 pKa = 10.41PAFDD526 pKa = 3.52QIGFQDD532 pKa = 5.58LITDD536 pKa = 4.21QMAFWDD542 pKa = 5.04TYY544 pKa = 10.39CVSGGIAPPVFKK556 pKa = 10.84SAGKK560 pKa = 8.98QPAWINYY567 pKa = 4.47QTEE570 pKa = 3.77IDD572 pKa = 3.52RR573 pKa = 11.84VFGNFAIQSAEE584 pKa = 3.08GWMVLDD590 pKa = 5.9RR591 pKa = 11.84NYY593 pKa = 11.57DD594 pKa = 3.13WDD596 pKa = 4.36AEE598 pKa = 4.13NHH600 pKa = 5.66TIKK603 pKa = 11.01DD604 pKa = 3.6LTTYY608 pKa = 10.51IDD610 pKa = 3.49PEE612 pKa = 4.2KK613 pKa = 10.66YY614 pKa = 10.31NGVFAYY620 pKa = 10.05RR621 pKa = 11.84ALDD624 pKa = 3.67AQNIWAQFKK633 pKa = 10.72VRR635 pKa = 11.84DD636 pKa = 3.47KK637 pKa = 11.15AIRR640 pKa = 11.84KK641 pKa = 8.17MSANQIPRR649 pKa = 11.84MM650 pKa = 4.0
Molecular weight: 73.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W775|A0A4P8W775_9VIRU Uncharacterized protein OS=Tortoise microvirus 85 OX=2583193 PE=4 SV=1
MM1 pKa = 7.74EE2 pKa = 5.23CRR4 pKa = 11.84KK5 pKa = 10.04QKK7 pKa = 10.71ARR9 pKa = 11.84DD10 pKa = 3.32WQVRR14 pKa = 11.84LLEE17 pKa = 4.3DD18 pKa = 3.46VRR20 pKa = 11.84HH21 pKa = 4.88NTNGKK26 pKa = 10.07FITLTLSNEE35 pKa = 4.1NFKK38 pKa = 11.01KK39 pKa = 10.42LAYY42 pKa = 8.55EE43 pKa = 4.14ANKK46 pKa = 8.5KK47 pKa = 6.93TKK49 pKa = 9.73KK50 pKa = 10.15HH51 pKa = 6.04PIGKK55 pKa = 9.29KK56 pKa = 9.89EE57 pKa = 3.97YY58 pKa = 9.06IDD60 pKa = 3.65KK61 pKa = 10.72NGKK64 pKa = 8.68IRR66 pKa = 11.84IRR68 pKa = 11.84YY69 pKa = 7.74KK70 pKa = 10.47YY71 pKa = 9.24KK72 pKa = 10.41TITEE76 pKa = 4.16QIEE79 pKa = 3.91LRR81 pKa = 11.84GYY83 pKa = 8.16EE84 pKa = 4.16TDD86 pKa = 3.88NEE88 pKa = 4.22IAKK91 pKa = 10.06LAVRR95 pKa = 11.84RR96 pKa = 11.84FLEE99 pKa = 4.33RR100 pKa = 11.84WRR102 pKa = 11.84KK103 pKa = 6.95TYY105 pKa = 10.51KK106 pKa = 10.0KK107 pKa = 10.23SLRR110 pKa = 11.84HH111 pKa = 5.01WLVTEE116 pKa = 4.84LGHH119 pKa = 7.04NGTEE123 pKa = 4.22NIHH126 pKa = 5.86LHH128 pKa = 5.91GIVWTNEE135 pKa = 3.75PIEE138 pKa = 4.85AIRR141 pKa = 11.84KK142 pKa = 7.59HH143 pKa = 5.55WDD145 pKa = 3.05YY146 pKa = 11.65GYY148 pKa = 8.78IWPRR152 pKa = 11.84KK153 pKa = 6.36EE154 pKa = 3.67HH155 pKa = 6.17EE156 pKa = 4.38KK157 pKa = 10.66VIKK160 pKa = 10.66NYY162 pKa = 10.34VNEE165 pKa = 3.96RR166 pKa = 11.84TINYY170 pKa = 4.73ITKK173 pKa = 9.86YY174 pKa = 10.32VSKK177 pKa = 10.81QDD179 pKa = 3.9FQHH182 pKa = 6.7KK183 pKa = 8.95EE184 pKa = 3.64YY185 pKa = 10.71KK186 pKa = 10.58AIILTSAGIGAGYY199 pKa = 6.74MTRR202 pKa = 11.84MDD204 pKa = 3.97SKK206 pKa = 10.51LNKK209 pKa = 10.16YY210 pKa = 10.13NGEE213 pKa = 4.1DD214 pKa = 3.24TKK216 pKa = 11.18DD217 pKa = 3.59YY218 pKa = 10.16YY219 pKa = 9.41VTRR222 pKa = 11.84TGHH225 pKa = 6.51KK226 pKa = 9.69IKK228 pKa = 10.63IPIYY232 pKa = 8.74WRR234 pKa = 11.84NKK236 pKa = 8.63IYY238 pKa = 10.63NDD240 pKa = 3.53EE241 pKa = 3.98EE242 pKa = 4.82RR243 pKa = 11.84EE244 pKa = 4.04RR245 pKa = 11.84LWLHH249 pKa = 6.41KK250 pKa = 10.28LDD252 pKa = 5.03EE253 pKa = 4.86KK254 pKa = 10.95IRR256 pKa = 11.84WIGKK260 pKa = 7.83EE261 pKa = 4.06KK262 pKa = 10.25IDD264 pKa = 3.41ISLNEE269 pKa = 3.81EE270 pKa = 3.96EE271 pKa = 4.86YY272 pKa = 11.23YY273 pKa = 9.29KK274 pKa = 10.27TLHH277 pKa = 7.15HH278 pKa = 6.12YY279 pKa = 8.7RR280 pKa = 11.84QKK282 pKa = 10.75NARR285 pKa = 11.84LGYY288 pKa = 11.06GNGTKK293 pKa = 10.06DD294 pKa = 3.3WNRR297 pKa = 11.84KK298 pKa = 6.67QYY300 pKa = 10.52EE301 pKa = 4.0EE302 pKa = 3.81QRR304 pKa = 11.84RR305 pKa = 11.84ILLQQEE311 pKa = 4.71RR312 pKa = 11.84IRR314 pKa = 11.84ATEE317 pKa = 4.16EE318 pKa = 3.41IEE320 pKa = 5.65AYY322 pKa = 9.76PEE324 pKa = 3.67WVYY327 pKa = 11.48NSSGGSCAEE336 pKa = 4.12SATCFQVDD344 pKa = 3.62QRR346 pKa = 11.84EE347 pKa = 4.34HH348 pKa = 5.71
MM1 pKa = 7.74EE2 pKa = 5.23CRR4 pKa = 11.84KK5 pKa = 10.04QKK7 pKa = 10.71ARR9 pKa = 11.84DD10 pKa = 3.32WQVRR14 pKa = 11.84LLEE17 pKa = 4.3DD18 pKa = 3.46VRR20 pKa = 11.84HH21 pKa = 4.88NTNGKK26 pKa = 10.07FITLTLSNEE35 pKa = 4.1NFKK38 pKa = 11.01KK39 pKa = 10.42LAYY42 pKa = 8.55EE43 pKa = 4.14ANKK46 pKa = 8.5KK47 pKa = 6.93TKK49 pKa = 9.73KK50 pKa = 10.15HH51 pKa = 6.04PIGKK55 pKa = 9.29KK56 pKa = 9.89EE57 pKa = 3.97YY58 pKa = 9.06IDD60 pKa = 3.65KK61 pKa = 10.72NGKK64 pKa = 8.68IRR66 pKa = 11.84IRR68 pKa = 11.84YY69 pKa = 7.74KK70 pKa = 10.47YY71 pKa = 9.24KK72 pKa = 10.41TITEE76 pKa = 4.16QIEE79 pKa = 3.91LRR81 pKa = 11.84GYY83 pKa = 8.16EE84 pKa = 4.16TDD86 pKa = 3.88NEE88 pKa = 4.22IAKK91 pKa = 10.06LAVRR95 pKa = 11.84RR96 pKa = 11.84FLEE99 pKa = 4.33RR100 pKa = 11.84WRR102 pKa = 11.84KK103 pKa = 6.95TYY105 pKa = 10.51KK106 pKa = 10.0KK107 pKa = 10.23SLRR110 pKa = 11.84HH111 pKa = 5.01WLVTEE116 pKa = 4.84LGHH119 pKa = 7.04NGTEE123 pKa = 4.22NIHH126 pKa = 5.86LHH128 pKa = 5.91GIVWTNEE135 pKa = 3.75PIEE138 pKa = 4.85AIRR141 pKa = 11.84KK142 pKa = 7.59HH143 pKa = 5.55WDD145 pKa = 3.05YY146 pKa = 11.65GYY148 pKa = 8.78IWPRR152 pKa = 11.84KK153 pKa = 6.36EE154 pKa = 3.67HH155 pKa = 6.17EE156 pKa = 4.38KK157 pKa = 10.66VIKK160 pKa = 10.66NYY162 pKa = 10.34VNEE165 pKa = 3.96RR166 pKa = 11.84TINYY170 pKa = 4.73ITKK173 pKa = 9.86YY174 pKa = 10.32VSKK177 pKa = 10.81QDD179 pKa = 3.9FQHH182 pKa = 6.7KK183 pKa = 8.95EE184 pKa = 3.64YY185 pKa = 10.71KK186 pKa = 10.58AIILTSAGIGAGYY199 pKa = 6.74MTRR202 pKa = 11.84MDD204 pKa = 3.97SKK206 pKa = 10.51LNKK209 pKa = 10.16YY210 pKa = 10.13NGEE213 pKa = 4.1DD214 pKa = 3.24TKK216 pKa = 11.18DD217 pKa = 3.59YY218 pKa = 10.16YY219 pKa = 9.41VTRR222 pKa = 11.84TGHH225 pKa = 6.51KK226 pKa = 9.69IKK228 pKa = 10.63IPIYY232 pKa = 8.74WRR234 pKa = 11.84NKK236 pKa = 8.63IYY238 pKa = 10.63NDD240 pKa = 3.53EE241 pKa = 3.98EE242 pKa = 4.82RR243 pKa = 11.84EE244 pKa = 4.04RR245 pKa = 11.84LWLHH249 pKa = 6.41KK250 pKa = 10.28LDD252 pKa = 5.03EE253 pKa = 4.86KK254 pKa = 10.95IRR256 pKa = 11.84WIGKK260 pKa = 7.83EE261 pKa = 4.06KK262 pKa = 10.25IDD264 pKa = 3.41ISLNEE269 pKa = 3.81EE270 pKa = 3.96EE271 pKa = 4.86YY272 pKa = 11.23YY273 pKa = 9.29KK274 pKa = 10.27TLHH277 pKa = 7.15HH278 pKa = 6.12YY279 pKa = 8.7RR280 pKa = 11.84QKK282 pKa = 10.75NARR285 pKa = 11.84LGYY288 pKa = 11.06GNGTKK293 pKa = 10.06DD294 pKa = 3.3WNRR297 pKa = 11.84KK298 pKa = 6.67QYY300 pKa = 10.52EE301 pKa = 4.0EE302 pKa = 3.81QRR304 pKa = 11.84RR305 pKa = 11.84ILLQQEE311 pKa = 4.71RR312 pKa = 11.84IRR314 pKa = 11.84ATEE317 pKa = 4.16EE318 pKa = 3.41IEE320 pKa = 5.65AYY322 pKa = 9.76PEE324 pKa = 3.67WVYY327 pKa = 11.48NSSGGSCAEE336 pKa = 4.12SATCFQVDD344 pKa = 3.62QRR346 pKa = 11.84EE347 pKa = 4.34HH348 pKa = 5.71
Molecular weight: 42.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1564 |
115 |
650 |
312.8 |
35.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.522 ± 1.221 | 0.32 ± 0.168 |
5.818 ± 0.886 | 7.801 ± 1.918 |
2.366 ± 0.747 | 7.353 ± 1.267 |
2.238 ± 0.697 | 8.376 ± 0.694 |
8.76 ± 1.733 | 7.289 ± 0.38 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.685 ± 0.757 | 6.394 ± 0.252 |
2.366 ± 0.751 | 5.179 ± 1.317 |
4.604 ± 1.139 | 4.476 ± 0.525 |
7.353 ± 0.829 | 3.772 ± 0.442 |
2.046 ± 0.375 | 4.284 ± 1.142 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
