
Photobacterium aphoticum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Photobacterium
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
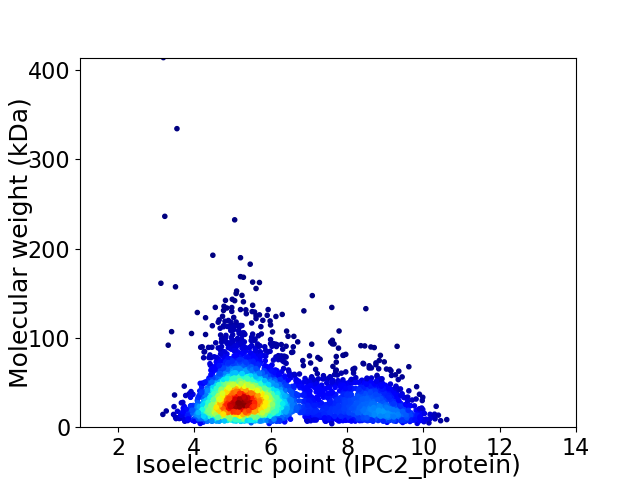
Virtual 2D-PAGE plot for 4349 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0J1GKN3|A0A0J1GKN3_9GAMM BOF domain-containing protein OS=Photobacterium aphoticum OX=754436 GN=ABT58_13535 PE=4 SV=1
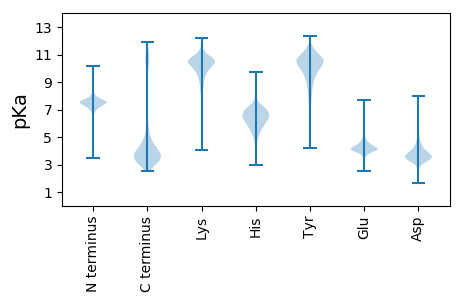
MM1 pKa = 7.65KK2 pKa = 9.0KK3 pKa = 8.22TLLALSVLAAGSAQAGINLYY23 pKa = 9.82DD24 pKa = 3.9ANGVTVDD31 pKa = 3.57LSGAVEE37 pKa = 4.0VQYY40 pKa = 10.89LQNIQDD46 pKa = 4.01KK47 pKa = 11.27NSDD50 pKa = 3.36ADD52 pKa = 3.86AGLHH56 pKa = 6.74LDD58 pKa = 5.68DD59 pKa = 6.75GDD61 pKa = 4.08LQLNTTIAVNEE72 pKa = 4.06QLNVVAGIGFAFEE85 pKa = 4.7KK86 pKa = 10.48RR87 pKa = 11.84DD88 pKa = 3.61VSNDD92 pKa = 3.48EE93 pKa = 3.88LWAGLSGDD101 pKa = 3.89FGTLTFGRR109 pKa = 11.84MLYY112 pKa = 10.13ISDD115 pKa = 3.89DD116 pKa = 3.51AGIGKK121 pKa = 9.92DD122 pKa = 3.55YY123 pKa = 11.05EE124 pKa = 4.44LGFGQVDD131 pKa = 4.1FVQTEE136 pKa = 4.07GNEE139 pKa = 4.09VIKK142 pKa = 10.83YY143 pKa = 10.23VFDD146 pKa = 3.59NGQFYY151 pKa = 10.75FGLSHH156 pKa = 7.62DD157 pKa = 5.12LDD159 pKa = 4.25TDD161 pKa = 3.72GSGASDD167 pKa = 3.71YY168 pKa = 11.4QNTDD172 pKa = 2.52GTVTDD177 pKa = 3.54VRR179 pKa = 11.84LGARR183 pKa = 11.84FADD186 pKa = 3.69LDD188 pKa = 3.69VRR190 pKa = 11.84GYY192 pKa = 11.02YY193 pKa = 8.13YY194 pKa = 10.6TGDD197 pKa = 4.56DD198 pKa = 3.52IQRR201 pKa = 11.84SADD204 pKa = 3.45QNGNPIFNSGTDD216 pKa = 2.9VDD218 pKa = 4.4AFNLEE223 pKa = 3.83AEE225 pKa = 4.43YY226 pKa = 11.8VMGQFAFAASYY237 pKa = 11.82GNFDD241 pKa = 3.5YY242 pKa = 11.62SNANGATYY250 pKa = 11.04SDD252 pKa = 3.2IDD254 pKa = 3.99VIEE257 pKa = 4.21VNGAYY262 pKa = 9.27TLGKK266 pKa = 8.35NTFALGYY273 pKa = 10.4NRR275 pKa = 11.84ADD277 pKa = 3.87DD278 pKa = 4.06SKK280 pKa = 11.63QDD282 pKa = 3.52VVTDD286 pKa = 3.41NVYY289 pKa = 11.46ANVTHH294 pKa = 6.15QLHH297 pKa = 6.73SNVKK301 pKa = 9.8VYY303 pKa = 11.26GEE305 pKa = 4.86LGWADD310 pKa = 3.41QDD312 pKa = 5.34GSDD315 pKa = 4.7LDD317 pKa = 4.03LGYY320 pKa = 10.93LVGMEE325 pKa = 4.19VKK327 pKa = 10.64FF328 pKa = 3.9
MM1 pKa = 7.65KK2 pKa = 9.0KK3 pKa = 8.22TLLALSVLAAGSAQAGINLYY23 pKa = 9.82DD24 pKa = 3.9ANGVTVDD31 pKa = 3.57LSGAVEE37 pKa = 4.0VQYY40 pKa = 10.89LQNIQDD46 pKa = 4.01KK47 pKa = 11.27NSDD50 pKa = 3.36ADD52 pKa = 3.86AGLHH56 pKa = 6.74LDD58 pKa = 5.68DD59 pKa = 6.75GDD61 pKa = 4.08LQLNTTIAVNEE72 pKa = 4.06QLNVVAGIGFAFEE85 pKa = 4.7KK86 pKa = 10.48RR87 pKa = 11.84DD88 pKa = 3.61VSNDD92 pKa = 3.48EE93 pKa = 3.88LWAGLSGDD101 pKa = 3.89FGTLTFGRR109 pKa = 11.84MLYY112 pKa = 10.13ISDD115 pKa = 3.89DD116 pKa = 3.51AGIGKK121 pKa = 9.92DD122 pKa = 3.55YY123 pKa = 11.05EE124 pKa = 4.44LGFGQVDD131 pKa = 4.1FVQTEE136 pKa = 4.07GNEE139 pKa = 4.09VIKK142 pKa = 10.83YY143 pKa = 10.23VFDD146 pKa = 3.59NGQFYY151 pKa = 10.75FGLSHH156 pKa = 7.62DD157 pKa = 5.12LDD159 pKa = 4.25TDD161 pKa = 3.72GSGASDD167 pKa = 3.71YY168 pKa = 11.4QNTDD172 pKa = 2.52GTVTDD177 pKa = 3.54VRR179 pKa = 11.84LGARR183 pKa = 11.84FADD186 pKa = 3.69LDD188 pKa = 3.69VRR190 pKa = 11.84GYY192 pKa = 11.02YY193 pKa = 8.13YY194 pKa = 10.6TGDD197 pKa = 4.56DD198 pKa = 3.52IQRR201 pKa = 11.84SADD204 pKa = 3.45QNGNPIFNSGTDD216 pKa = 2.9VDD218 pKa = 4.4AFNLEE223 pKa = 3.83AEE225 pKa = 4.43YY226 pKa = 11.8VMGQFAFAASYY237 pKa = 11.82GNFDD241 pKa = 3.5YY242 pKa = 11.62SNANGATYY250 pKa = 11.04SDD252 pKa = 3.2IDD254 pKa = 3.99VIEE257 pKa = 4.21VNGAYY262 pKa = 9.27TLGKK266 pKa = 8.35NTFALGYY273 pKa = 10.4NRR275 pKa = 11.84ADD277 pKa = 3.87DD278 pKa = 4.06SKK280 pKa = 11.63QDD282 pKa = 3.52VVTDD286 pKa = 3.41NVYY289 pKa = 11.46ANVTHH294 pKa = 6.15QLHH297 pKa = 6.73SNVKK301 pKa = 9.8VYY303 pKa = 11.26GEE305 pKa = 4.86LGWADD310 pKa = 3.41QDD312 pKa = 5.34GSDD315 pKa = 4.7LDD317 pKa = 4.03LGYY320 pKa = 10.93LVGMEE325 pKa = 4.19VKK327 pKa = 10.64FF328 pKa = 3.9
Molecular weight: 35.47 kDa
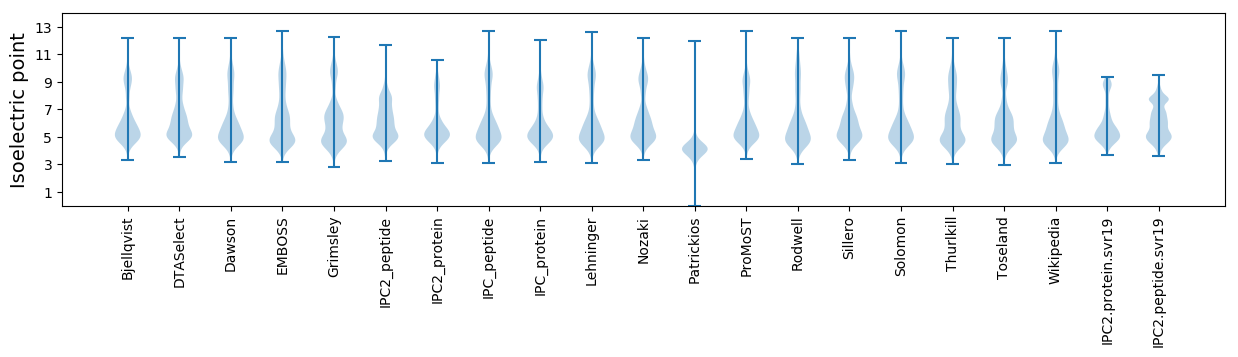
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0J1GN04|A0A0J1GN04_9GAMM Uncharacterized protein OS=Photobacterium aphoticum OX=754436 GN=ABT58_10735 PE=4 SV=1
MM1 pKa = 7.34PRR3 pKa = 11.84HH4 pKa = 5.23VIKK7 pKa = 10.68RR8 pKa = 11.84FMPSHH13 pKa = 6.21EE14 pKa = 4.35LIKK17 pKa = 10.35RR18 pKa = 11.84QKK20 pKa = 9.84ALKK23 pKa = 10.18IFGNVLYY30 pKa = 10.72NPNLWCLNRR39 pKa = 11.84RR40 pKa = 11.84SASGAFAVGLFMAFVPLPSQMIMAAGLAILFSVNLPLSVCLVWISNPITMPVLFYY95 pKa = 10.95GAYY98 pKa = 9.47KK99 pKa = 10.2VGAWLMSTPHH109 pKa = 5.52QAFHH113 pKa = 7.45FEE115 pKa = 4.26LSWEE119 pKa = 4.2FLLHH123 pKa = 6.03QMNQIGPPFLLGCFVTGVTCALIGYY148 pKa = 8.25FGIRR152 pKa = 11.84GLWRR156 pKa = 11.84YY157 pKa = 9.89SVVRR161 pKa = 11.84SWQKK165 pKa = 10.2RR166 pKa = 11.84KK167 pKa = 9.73FRR169 pKa = 11.84VLRR172 pKa = 4.07
MM1 pKa = 7.34PRR3 pKa = 11.84HH4 pKa = 5.23VIKK7 pKa = 10.68RR8 pKa = 11.84FMPSHH13 pKa = 6.21EE14 pKa = 4.35LIKK17 pKa = 10.35RR18 pKa = 11.84QKK20 pKa = 9.84ALKK23 pKa = 10.18IFGNVLYY30 pKa = 10.72NPNLWCLNRR39 pKa = 11.84RR40 pKa = 11.84SASGAFAVGLFMAFVPLPSQMIMAAGLAILFSVNLPLSVCLVWISNPITMPVLFYY95 pKa = 10.95GAYY98 pKa = 9.47KK99 pKa = 10.2VGAWLMSTPHH109 pKa = 5.52QAFHH113 pKa = 7.45FEE115 pKa = 4.26LSWEE119 pKa = 4.2FLLHH123 pKa = 6.03QMNQIGPPFLLGCFVTGVTCALIGYY148 pKa = 8.25FGIRR152 pKa = 11.84GLWRR156 pKa = 11.84YY157 pKa = 9.89SVVRR161 pKa = 11.84SWQKK165 pKa = 10.2RR166 pKa = 11.84KK167 pKa = 9.73FRR169 pKa = 11.84VLRR172 pKa = 4.07
Molecular weight: 19.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1394034 |
35 |
4031 |
320.5 |
35.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.631 ± 0.047 | 1.084 ± 0.014 |
5.706 ± 0.041 | 5.851 ± 0.035 |
3.985 ± 0.026 | 6.969 ± 0.04 |
2.474 ± 0.024 | 5.923 ± 0.03 |
4.523 ± 0.035 | 10.285 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.878 ± 0.019 | 3.863 ± 0.026 |
4.22 ± 0.022 | 4.747 ± 0.03 |
4.724 ± 0.031 | 5.93 ± 0.03 |
5.682 ± 0.036 | 7.132 ± 0.033 |
1.319 ± 0.017 | 3.072 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
