
Lake Sarah-associated circular virus-50
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
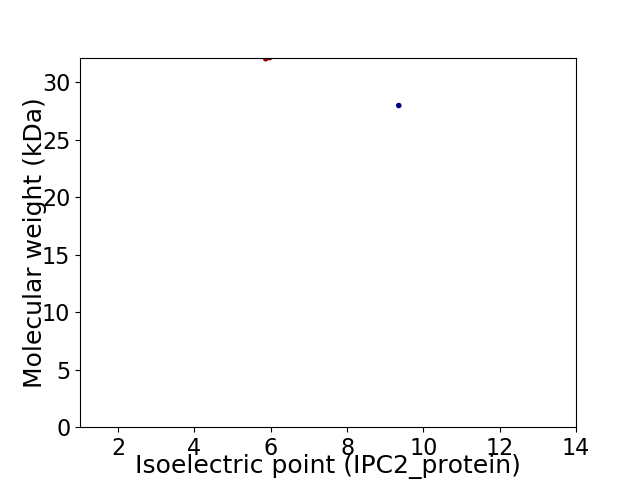
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126G9T2|A0A126G9T2_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-50 OX=1685780 PE=4 SV=1
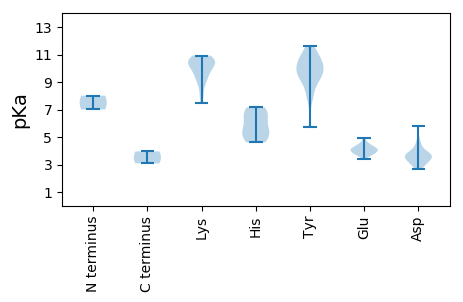
MM1 pKa = 7.0VKK3 pKa = 10.3SRR5 pKa = 11.84GWCYY9 pKa = 9.01TANNYY14 pKa = 7.74TKK16 pKa = 10.64EE17 pKa = 4.21EE18 pKa = 4.27YY19 pKa = 10.19EE20 pKa = 4.13SLEE23 pKa = 4.34KK24 pKa = 10.67YY25 pKa = 10.31DD26 pKa = 3.84CGYY29 pKa = 10.8HH30 pKa = 7.15VMGKK34 pKa = 9.39EE35 pKa = 3.97VGEE38 pKa = 4.05QGTPHH43 pKa = 5.46IQGYY47 pKa = 9.8IEE49 pKa = 3.91FTNAKK54 pKa = 10.2RR55 pKa = 11.84FDD57 pKa = 3.76TLHH60 pKa = 6.83ADD62 pKa = 3.77FPKK65 pKa = 10.38IHH67 pKa = 6.46WEE69 pKa = 3.93PRR71 pKa = 11.84KK72 pKa = 8.82GTQQQAIDD80 pKa = 3.71YY81 pKa = 9.0CMKK84 pKa = 10.7GGDD87 pKa = 3.72FVEE90 pKa = 4.92IGTKK94 pKa = 9.74KK95 pKa = 10.65DD96 pKa = 2.97QGKK99 pKa = 8.01RR100 pKa = 11.84TDD102 pKa = 3.18IEE104 pKa = 4.45AVIKK108 pKa = 10.32DD109 pKa = 3.4INDD112 pKa = 3.44KK113 pKa = 10.84VFSPNNHH120 pKa = 4.61AVAYY124 pKa = 9.17IKK126 pKa = 10.05YY127 pKa = 10.12AKK129 pKa = 10.76GIDD132 pKa = 3.63CYY134 pKa = 11.64ANYY137 pKa = 10.53KK138 pKa = 9.95LLPRR142 pKa = 11.84TVKK145 pKa = 9.22PTVEE149 pKa = 3.41WRR151 pKa = 11.84YY152 pKa = 10.73GLTGSGKK159 pKa = 7.5TRR161 pKa = 11.84EE162 pKa = 4.0PYY164 pKa = 9.42EE165 pKa = 3.72RR166 pKa = 11.84HH167 pKa = 5.93INSVYY172 pKa = 10.63IKK174 pKa = 10.54DD175 pKa = 3.42GTMWWNNYY183 pKa = 5.7TQQEE187 pKa = 4.1AVIIDD192 pKa = 4.55DD193 pKa = 5.79FDD195 pKa = 3.85GHH197 pKa = 5.22WPYY200 pKa = 11.46RR201 pKa = 11.84DD202 pKa = 3.7FLRR205 pKa = 11.84LLDD208 pKa = 4.28RR209 pKa = 11.84YY210 pKa = 9.52PYY212 pKa = 9.34QGQYY216 pKa = 9.58KK217 pKa = 9.59GGYY220 pKa = 8.29VNISSAYY227 pKa = 9.45IYY229 pKa = 8.14ITCEE233 pKa = 3.79FSPEE237 pKa = 4.18TFWRR241 pKa = 11.84GNEE244 pKa = 3.65LAQVIRR250 pKa = 11.84RR251 pKa = 11.84IDD253 pKa = 3.75TITLCRR259 pKa = 11.84INEE262 pKa = 4.23PVPVMVIDD270 pKa = 4.83DD271 pKa = 4.26LVDD274 pKa = 3.08
MM1 pKa = 7.0VKK3 pKa = 10.3SRR5 pKa = 11.84GWCYY9 pKa = 9.01TANNYY14 pKa = 7.74TKK16 pKa = 10.64EE17 pKa = 4.21EE18 pKa = 4.27YY19 pKa = 10.19EE20 pKa = 4.13SLEE23 pKa = 4.34KK24 pKa = 10.67YY25 pKa = 10.31DD26 pKa = 3.84CGYY29 pKa = 10.8HH30 pKa = 7.15VMGKK34 pKa = 9.39EE35 pKa = 3.97VGEE38 pKa = 4.05QGTPHH43 pKa = 5.46IQGYY47 pKa = 9.8IEE49 pKa = 3.91FTNAKK54 pKa = 10.2RR55 pKa = 11.84FDD57 pKa = 3.76TLHH60 pKa = 6.83ADD62 pKa = 3.77FPKK65 pKa = 10.38IHH67 pKa = 6.46WEE69 pKa = 3.93PRR71 pKa = 11.84KK72 pKa = 8.82GTQQQAIDD80 pKa = 3.71YY81 pKa = 9.0CMKK84 pKa = 10.7GGDD87 pKa = 3.72FVEE90 pKa = 4.92IGTKK94 pKa = 9.74KK95 pKa = 10.65DD96 pKa = 2.97QGKK99 pKa = 8.01RR100 pKa = 11.84TDD102 pKa = 3.18IEE104 pKa = 4.45AVIKK108 pKa = 10.32DD109 pKa = 3.4INDD112 pKa = 3.44KK113 pKa = 10.84VFSPNNHH120 pKa = 4.61AVAYY124 pKa = 9.17IKK126 pKa = 10.05YY127 pKa = 10.12AKK129 pKa = 10.76GIDD132 pKa = 3.63CYY134 pKa = 11.64ANYY137 pKa = 10.53KK138 pKa = 9.95LLPRR142 pKa = 11.84TVKK145 pKa = 9.22PTVEE149 pKa = 3.41WRR151 pKa = 11.84YY152 pKa = 10.73GLTGSGKK159 pKa = 7.5TRR161 pKa = 11.84EE162 pKa = 4.0PYY164 pKa = 9.42EE165 pKa = 3.72RR166 pKa = 11.84HH167 pKa = 5.93INSVYY172 pKa = 10.63IKK174 pKa = 10.54DD175 pKa = 3.42GTMWWNNYY183 pKa = 5.7TQQEE187 pKa = 4.1AVIIDD192 pKa = 4.55DD193 pKa = 5.79FDD195 pKa = 3.85GHH197 pKa = 5.22WPYY200 pKa = 11.46RR201 pKa = 11.84DD202 pKa = 3.7FLRR205 pKa = 11.84LLDD208 pKa = 4.28RR209 pKa = 11.84YY210 pKa = 9.52PYY212 pKa = 9.34QGQYY216 pKa = 9.58KK217 pKa = 9.59GGYY220 pKa = 8.29VNISSAYY227 pKa = 9.45IYY229 pKa = 8.14ITCEE233 pKa = 3.79FSPEE237 pKa = 4.18TFWRR241 pKa = 11.84GNEE244 pKa = 3.65LAQVIRR250 pKa = 11.84RR251 pKa = 11.84IDD253 pKa = 3.75TITLCRR259 pKa = 11.84INEE262 pKa = 4.23PVPVMVIDD270 pKa = 4.83DD271 pKa = 4.26LVDD274 pKa = 3.08
Molecular weight: 32.05 kDa
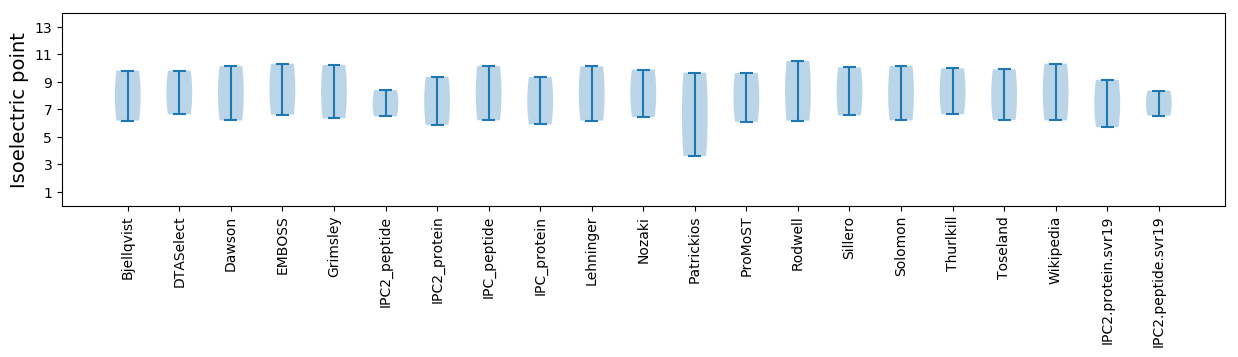
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G9T2|A0A126G9T2_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-50 OX=1685780 PE=4 SV=1
MM1 pKa = 8.01PYY3 pKa = 11.14AKK5 pKa = 9.25MVKK8 pKa = 9.9GRR10 pKa = 11.84VAARR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 5.32PSKK20 pKa = 10.8KK21 pKa = 9.16YY22 pKa = 9.86VKK24 pKa = 8.2PTKK27 pKa = 10.51SFTEE31 pKa = 4.06KK32 pKa = 9.97VVKK35 pKa = 10.35VINRR39 pKa = 11.84RR40 pKa = 11.84LDD42 pKa = 3.6VKK44 pKa = 10.89HH45 pKa = 6.76LDD47 pKa = 3.26AGYY50 pKa = 9.67GATAIVLRR58 pKa = 11.84NNTGPVAPTIVEE70 pKa = 4.51LAQSLIGTIGQGPGEE85 pKa = 4.19YY86 pKa = 9.9EE87 pKa = 3.73RR88 pKa = 11.84LGNDD92 pKa = 2.83IRR94 pKa = 11.84VKK96 pKa = 9.68SCIMRR101 pKa = 11.84GTLYY105 pKa = 9.33NTNATQRR112 pKa = 11.84PFYY115 pKa = 10.72IKK117 pKa = 10.22MFLLKK122 pKa = 10.69NKK124 pKa = 9.26LVPAADD130 pKa = 3.55TFNNLFEE137 pKa = 4.59GTTGNPTNQLSDD149 pKa = 3.08LTRR152 pKa = 11.84RR153 pKa = 11.84VNTDD157 pKa = 2.64EE158 pKa = 3.79YY159 pKa = 10.57TYY161 pKa = 10.54IASRR165 pKa = 11.84TFKK168 pKa = 10.52LGPSTATTNPNNDD181 pKa = 2.98FKK183 pKa = 10.92MSQFWRR189 pKa = 11.84IDD191 pKa = 3.27LTKK194 pKa = 10.38HH195 pKa = 4.73MKK197 pKa = 8.73IVKK200 pKa = 10.06YY201 pKa = 10.05PIDD204 pKa = 3.91GNGTVSQPASLFLVFVGAYY223 pKa = 10.04ADD225 pKa = 3.52NTAITFGAYY234 pKa = 7.96TGPAVSVSYY243 pKa = 11.16EE244 pKa = 3.8MTLDD248 pKa = 3.54FTDD251 pKa = 3.23AA252 pKa = 3.95
MM1 pKa = 8.01PYY3 pKa = 11.14AKK5 pKa = 9.25MVKK8 pKa = 9.9GRR10 pKa = 11.84VAARR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 5.32PSKK20 pKa = 10.8KK21 pKa = 9.16YY22 pKa = 9.86VKK24 pKa = 8.2PTKK27 pKa = 10.51SFTEE31 pKa = 4.06KK32 pKa = 9.97VVKK35 pKa = 10.35VINRR39 pKa = 11.84RR40 pKa = 11.84LDD42 pKa = 3.6VKK44 pKa = 10.89HH45 pKa = 6.76LDD47 pKa = 3.26AGYY50 pKa = 9.67GATAIVLRR58 pKa = 11.84NNTGPVAPTIVEE70 pKa = 4.51LAQSLIGTIGQGPGEE85 pKa = 4.19YY86 pKa = 9.9EE87 pKa = 3.73RR88 pKa = 11.84LGNDD92 pKa = 2.83IRR94 pKa = 11.84VKK96 pKa = 9.68SCIMRR101 pKa = 11.84GTLYY105 pKa = 9.33NTNATQRR112 pKa = 11.84PFYY115 pKa = 10.72IKK117 pKa = 10.22MFLLKK122 pKa = 10.69NKK124 pKa = 9.26LVPAADD130 pKa = 3.55TFNNLFEE137 pKa = 4.59GTTGNPTNQLSDD149 pKa = 3.08LTRR152 pKa = 11.84RR153 pKa = 11.84VNTDD157 pKa = 2.64EE158 pKa = 3.79YY159 pKa = 10.57TYY161 pKa = 10.54IASRR165 pKa = 11.84TFKK168 pKa = 10.52LGPSTATTNPNNDD181 pKa = 2.98FKK183 pKa = 10.92MSQFWRR189 pKa = 11.84IDD191 pKa = 3.27LTKK194 pKa = 10.38HH195 pKa = 4.73MKK197 pKa = 8.73IVKK200 pKa = 10.06YY201 pKa = 10.05PIDD204 pKa = 3.91GNGTVSQPASLFLVFVGAYY223 pKa = 10.04ADD225 pKa = 3.52NTAITFGAYY234 pKa = 7.96TGPAVSVSYY243 pKa = 11.16EE244 pKa = 3.8MTLDD248 pKa = 3.54FTDD251 pKa = 3.23AA252 pKa = 3.95
Molecular weight: 27.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
526 |
252 |
274 |
263.0 |
30.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.084 ± 1.191 | 1.331 ± 0.601 |
6.084 ± 0.85 | 4.753 ± 1.27 |
3.992 ± 0.495 | 7.414 ± 0.175 |
1.901 ± 0.457 | 6.654 ± 0.961 |
7.224 ± 0.052 | 5.513 ± 1.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.281 ± 0.319 | 5.703 ± 0.67 |
4.943 ± 0.394 | 3.232 ± 0.547 |
5.703 ± 0.16 | 3.802 ± 0.617 |
8.555 ± 1.388 | 6.844 ± 0.447 |
1.521 ± 0.723 | 6.464 ± 1.094 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
