
Miniopterus schreibersii papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyotaupapillomavirus; Dyotaupapillomavirus 1
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
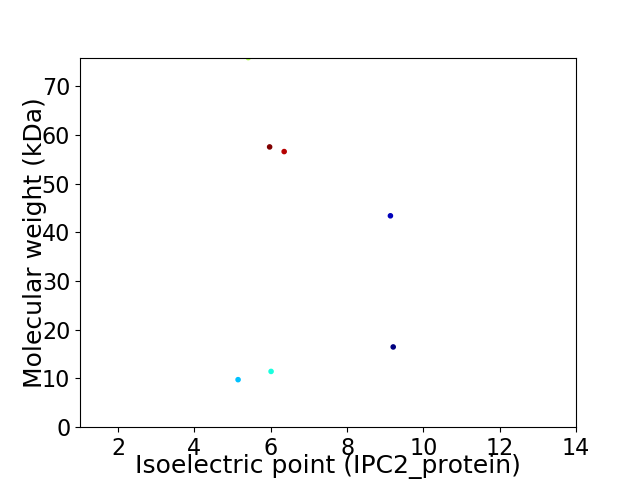
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J9R1I3|J9R1I3_9PAPI Major capsid protein L1 OS=Miniopterus schreibersii papillomavirus 1 OX=1195364 GN=L1 PE=3 SV=1
MM1 pKa = 7.71LYY3 pKa = 10.96DD4 pKa = 4.93NIPGYY9 pKa = 6.59TTLDD13 pKa = 3.59FVEE16 pKa = 4.78SVQAAATTNSQGQLVVLSPCSIGASCCCCGYY47 pKa = 10.73VVVLHH52 pKa = 6.6FVATEE57 pKa = 3.74DD58 pKa = 3.61AAIVVDD64 pKa = 4.3NLITTARR71 pKa = 11.84LNLFCTRR78 pKa = 11.84CVADD82 pKa = 4.45LKK84 pKa = 10.45RR85 pKa = 11.84ARR87 pKa = 11.84DD88 pKa = 3.72GRR90 pKa = 11.84QQ91 pKa = 2.65
MM1 pKa = 7.71LYY3 pKa = 10.96DD4 pKa = 4.93NIPGYY9 pKa = 6.59TTLDD13 pKa = 3.59FVEE16 pKa = 4.78SVQAAATTNSQGQLVVLSPCSIGASCCCCGYY47 pKa = 10.73VVVLHH52 pKa = 6.6FVATEE57 pKa = 3.74DD58 pKa = 3.61AAIVVDD64 pKa = 4.3NLITTARR71 pKa = 11.84LNLFCTRR78 pKa = 11.84CVADD82 pKa = 4.45LKK84 pKa = 10.45RR85 pKa = 11.84ARR87 pKa = 11.84DD88 pKa = 3.72GRR90 pKa = 11.84QQ91 pKa = 2.65
Molecular weight: 9.76 kDa
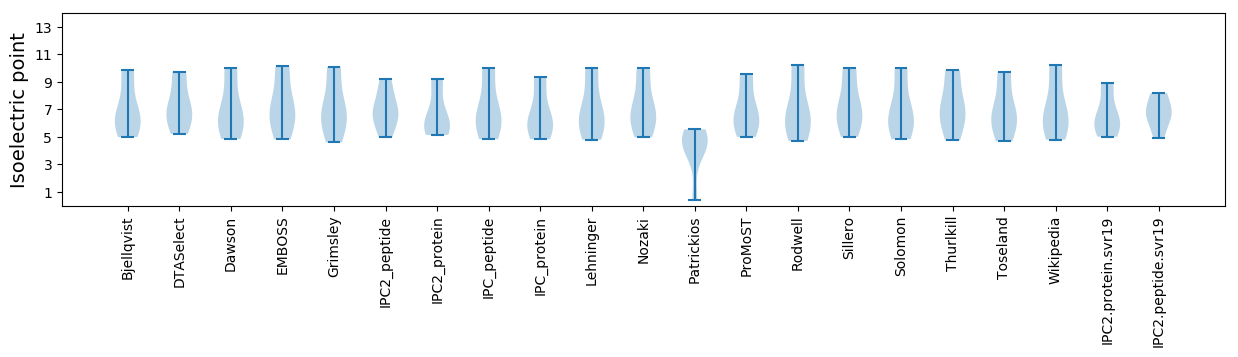
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J9QY83|J9QY83_9PAPI Replication protein E1 OS=Miniopterus schreibersii papillomavirus 1 OX=1195364 GN=E1 PE=3 SV=1
MM1 pKa = 7.08NQRR4 pKa = 11.84TDD6 pKa = 3.31PLTARR11 pKa = 11.84LEE13 pKa = 4.37SVQEE17 pKa = 4.17EE18 pKa = 4.11IFEE21 pKa = 5.05LIEE24 pKa = 4.06EE25 pKa = 4.33SSNRR29 pKa = 11.84LEE31 pKa = 3.98DD32 pKa = 3.64QIRR35 pKa = 11.84YY36 pKa = 8.43WEE38 pKa = 4.87LIRR41 pKa = 11.84KK42 pKa = 7.27EE43 pKa = 3.87QALLFLARR51 pKa = 11.84RR52 pKa = 11.84EE53 pKa = 4.02RR54 pKa = 11.84ITRR57 pKa = 11.84IGAEE61 pKa = 4.22VVPPLSVSEE70 pKa = 4.29TRR72 pKa = 11.84AKK74 pKa = 10.35QAIQMSLLLTSLNKK88 pKa = 9.89SHH90 pKa = 6.71YY91 pKa = 10.21KK92 pKa = 9.87NEE94 pKa = 3.83PWTMQEE100 pKa = 3.78TSRR103 pKa = 11.84EE104 pKa = 3.88RR105 pKa = 11.84MLAPPKK111 pKa = 9.72YY112 pKa = 8.11TFKK115 pKa = 11.1KK116 pKa = 9.82AGKK119 pKa = 9.28SVDD122 pKa = 3.46IIFDD126 pKa = 3.88GNIEE130 pKa = 4.04NSVRR134 pKa = 11.84EE135 pKa = 4.24TMWGFIYY142 pKa = 10.89YY143 pKa = 9.65QDD145 pKa = 5.83SDD147 pKa = 5.61DD148 pKa = 4.07EE149 pKa = 4.43WQKK152 pKa = 10.98QPGEE156 pKa = 4.17IDD158 pKa = 3.63DD159 pKa = 3.68QGLFYY164 pKa = 10.56RR165 pKa = 11.84DD166 pKa = 3.2YY167 pKa = 11.27DD168 pKa = 3.68NEE170 pKa = 4.12KK171 pKa = 10.1IYY173 pKa = 11.21YY174 pKa = 9.98VDD176 pKa = 4.28FKK178 pKa = 11.2EE179 pKa = 4.96LAAKK183 pKa = 9.91YY184 pKa = 9.93SKK186 pKa = 10.19EE187 pKa = 3.52GRR189 pKa = 11.84YY190 pKa = 9.41KK191 pKa = 10.97VIVDD195 pKa = 4.07SKK197 pKa = 8.67TVADD201 pKa = 3.88VVVSHH206 pKa = 7.19LDD208 pKa = 3.35NSEE211 pKa = 4.13SPVRR215 pKa = 11.84TKK217 pKa = 10.6AAPAGRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84ISGGRR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84PRR234 pKa = 11.84GRR236 pKa = 11.84SSSHH240 pKa = 5.24STPASTSSSSTPTRR254 pKa = 11.84SRR256 pKa = 11.84SRR258 pKa = 11.84PRR260 pKa = 11.84RR261 pKa = 11.84AYY263 pKa = 9.25TGAAPSPRR271 pKa = 11.84EE272 pKa = 3.81VGTQHH277 pKa = 7.19ASVTGPSGSRR287 pKa = 11.84LRR289 pKa = 11.84RR290 pKa = 11.84LLQEE294 pKa = 4.59ARR296 pKa = 11.84DD297 pKa = 3.98PPAVILSGPANTVKK311 pKa = 10.63CLRR314 pKa = 11.84FRR316 pKa = 11.84CRR318 pKa = 11.84HH319 pKa = 5.99RR320 pKa = 11.84YY321 pKa = 9.02NEE323 pKa = 4.03YY324 pKa = 9.98FDD326 pKa = 5.63KK327 pKa = 10.92ISTTWWWTGDD337 pKa = 2.81GRR339 pKa = 11.84TRR341 pKa = 11.84TGDD344 pKa = 2.93ASMLVSFQSEE354 pKa = 4.24SQRR357 pKa = 11.84TSFLNTVPVPSSVTVSAANLFLRR380 pKa = 4.84
MM1 pKa = 7.08NQRR4 pKa = 11.84TDD6 pKa = 3.31PLTARR11 pKa = 11.84LEE13 pKa = 4.37SVQEE17 pKa = 4.17EE18 pKa = 4.11IFEE21 pKa = 5.05LIEE24 pKa = 4.06EE25 pKa = 4.33SSNRR29 pKa = 11.84LEE31 pKa = 3.98DD32 pKa = 3.64QIRR35 pKa = 11.84YY36 pKa = 8.43WEE38 pKa = 4.87LIRR41 pKa = 11.84KK42 pKa = 7.27EE43 pKa = 3.87QALLFLARR51 pKa = 11.84RR52 pKa = 11.84EE53 pKa = 4.02RR54 pKa = 11.84ITRR57 pKa = 11.84IGAEE61 pKa = 4.22VVPPLSVSEE70 pKa = 4.29TRR72 pKa = 11.84AKK74 pKa = 10.35QAIQMSLLLTSLNKK88 pKa = 9.89SHH90 pKa = 6.71YY91 pKa = 10.21KK92 pKa = 9.87NEE94 pKa = 3.83PWTMQEE100 pKa = 3.78TSRR103 pKa = 11.84EE104 pKa = 3.88RR105 pKa = 11.84MLAPPKK111 pKa = 9.72YY112 pKa = 8.11TFKK115 pKa = 11.1KK116 pKa = 9.82AGKK119 pKa = 9.28SVDD122 pKa = 3.46IIFDD126 pKa = 3.88GNIEE130 pKa = 4.04NSVRR134 pKa = 11.84EE135 pKa = 4.24TMWGFIYY142 pKa = 10.89YY143 pKa = 9.65QDD145 pKa = 5.83SDD147 pKa = 5.61DD148 pKa = 4.07EE149 pKa = 4.43WQKK152 pKa = 10.98QPGEE156 pKa = 4.17IDD158 pKa = 3.63DD159 pKa = 3.68QGLFYY164 pKa = 10.56RR165 pKa = 11.84DD166 pKa = 3.2YY167 pKa = 11.27DD168 pKa = 3.68NEE170 pKa = 4.12KK171 pKa = 10.1IYY173 pKa = 11.21YY174 pKa = 9.98VDD176 pKa = 4.28FKK178 pKa = 11.2EE179 pKa = 4.96LAAKK183 pKa = 9.91YY184 pKa = 9.93SKK186 pKa = 10.19EE187 pKa = 3.52GRR189 pKa = 11.84YY190 pKa = 9.41KK191 pKa = 10.97VIVDD195 pKa = 4.07SKK197 pKa = 8.67TVADD201 pKa = 3.88VVVSHH206 pKa = 7.19LDD208 pKa = 3.35NSEE211 pKa = 4.13SPVRR215 pKa = 11.84TKK217 pKa = 10.6AAPAGRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84ISGGRR230 pKa = 11.84RR231 pKa = 11.84RR232 pKa = 11.84PRR234 pKa = 11.84GRR236 pKa = 11.84SSSHH240 pKa = 5.24STPASTSSSSTPTRR254 pKa = 11.84SRR256 pKa = 11.84SRR258 pKa = 11.84PRR260 pKa = 11.84RR261 pKa = 11.84AYY263 pKa = 9.25TGAAPSPRR271 pKa = 11.84EE272 pKa = 3.81VGTQHH277 pKa = 7.19ASVTGPSGSRR287 pKa = 11.84LRR289 pKa = 11.84RR290 pKa = 11.84LLQEE294 pKa = 4.59ARR296 pKa = 11.84DD297 pKa = 3.98PPAVILSGPANTVKK311 pKa = 10.63CLRR314 pKa = 11.84FRR316 pKa = 11.84CRR318 pKa = 11.84HH319 pKa = 5.99RR320 pKa = 11.84YY321 pKa = 9.02NEE323 pKa = 4.03YY324 pKa = 9.98FDD326 pKa = 5.63KK327 pKa = 10.92ISTTWWWTGDD337 pKa = 2.81GRR339 pKa = 11.84TRR341 pKa = 11.84TGDD344 pKa = 2.93ASMLVSFQSEE354 pKa = 4.24SQRR357 pKa = 11.84TSFLNTVPVPSSVTVSAANLFLRR380 pKa = 4.84
Molecular weight: 43.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2412 |
91 |
669 |
344.6 |
38.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.965 ± 0.364 | 1.866 ± 0.711 |
5.846 ± 0.488 | 6.551 ± 0.505 |
4.063 ± 0.315 | 6.385 ± 0.799 |
2.612 ± 0.425 | 4.685 ± 0.331 |
4.809 ± 0.747 | 8.831 ± 0.821 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.824 ± 0.232 | 3.483 ± 0.405 |
6.302 ± 0.806 | 4.063 ± 0.24 |
7.67 ± 1.049 | 7.09 ± 0.902 |
6.965 ± 0.491 | 5.97 ± 0.778 |
1.119 ± 0.285 | 2.902 ± 0.448 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
