
Piscibacillus halophilus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Piscibacillus
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
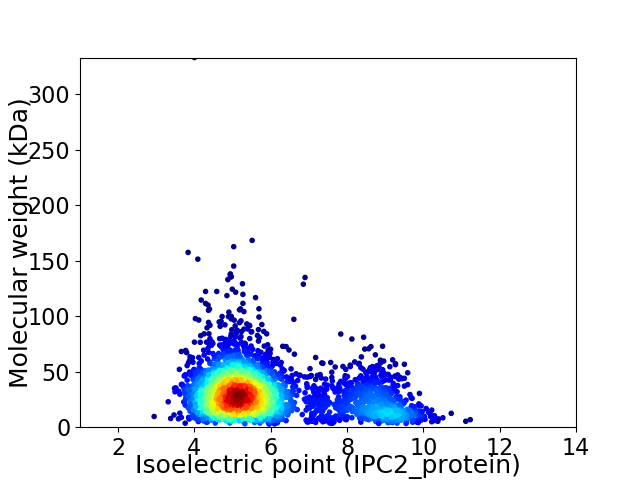
Virtual 2D-PAGE plot for 3140 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9I7Y6|A0A1H9I7Y6_9BACI 3-isopropylmalate dehydratase small subunit OS=Piscibacillus halophilus OX=571933 GN=leuD PE=3 SV=1
MM1 pKa = 7.52KK2 pKa = 10.5LKK4 pKa = 10.68SFWVALLLGLVLVLSACSGGDD25 pKa = 3.35SDD27 pKa = 6.36DD28 pKa = 4.62EE29 pKa = 4.39SAEE32 pKa = 4.34SNGEE36 pKa = 4.12KK37 pKa = 8.98ITIFQTKK44 pKa = 9.59VEE46 pKa = 4.09ISEE49 pKa = 4.1QMEE52 pKa = 4.22EE53 pKa = 3.6LAEE56 pKa = 4.25IYY58 pKa = 10.45EE59 pKa = 4.33EE60 pKa = 4.16EE61 pKa = 4.18TGVEE65 pKa = 4.15VEE67 pKa = 4.2VWGTTGDD74 pKa = 4.73DD75 pKa = 3.85YY76 pKa = 11.41FQQLQTRR83 pKa = 11.84LNSNQGPSILSLQDD97 pKa = 3.06YY98 pKa = 10.44KK99 pKa = 11.02KK100 pKa = 10.99AEE102 pKa = 4.23NIKK105 pKa = 10.72SYY107 pKa = 10.82LHH109 pKa = 6.77DD110 pKa = 4.2LSGEE114 pKa = 4.03EE115 pKa = 4.68FIDD118 pKa = 4.47NIAPNMAVEE127 pKa = 4.09IDD129 pKa = 4.01GEE131 pKa = 4.88VVGIPYY137 pKa = 10.19GVEE140 pKa = 3.97GFGLVYY146 pKa = 10.9NKK148 pKa = 10.72DD149 pKa = 3.62LVSEE153 pKa = 4.45EE154 pKa = 5.26DD155 pKa = 3.32IQDD158 pKa = 3.21YY159 pKa = 11.3DD160 pKa = 3.96SFVNTLEE167 pKa = 4.03RR168 pKa = 11.84LSNEE172 pKa = 3.89GVDD175 pKa = 3.96PLTLSSEE182 pKa = 4.29AYY184 pKa = 10.13FLIGHH189 pKa = 6.36MSNYY193 pKa = 9.28PFSLQEE199 pKa = 3.81DD200 pKa = 3.75HH201 pKa = 7.47FEE203 pKa = 4.69YY204 pKa = 10.08IDD206 pKa = 3.53QLSEE210 pKa = 4.48GEE212 pKa = 4.29VTMTEE217 pKa = 3.9TPEE220 pKa = 3.74FQEE223 pKa = 3.94FGKK226 pKa = 10.34FLEE229 pKa = 5.09AIRR232 pKa = 11.84EE233 pKa = 4.02YY234 pKa = 11.09SPNPMDD240 pKa = 3.41YY241 pKa = 10.29TYY243 pKa = 10.81DD244 pKa = 3.43EE245 pKa = 4.22QMGDD249 pKa = 3.29FATGKK254 pKa = 7.99TAMVHH259 pKa = 4.85QGNWAMGMLADD270 pKa = 3.89YY271 pKa = 10.66DD272 pKa = 4.14YY273 pKa = 11.39DD274 pKa = 4.31FEE276 pKa = 5.21VGMMPFPLAGNDD288 pKa = 3.36KK289 pKa = 10.58LAVGVGANWAVNGTKK304 pKa = 10.42DD305 pKa = 3.32EE306 pKa = 4.54AEE308 pKa = 3.91IEE310 pKa = 4.18AAVDD314 pKa = 3.75FLEE317 pKa = 4.2WLHH320 pKa = 6.0TSEE323 pKa = 3.78TGQRR327 pKa = 11.84FVVEE331 pKa = 4.03EE332 pKa = 4.28FGFVPAMTNIEE343 pKa = 4.28ANDD346 pKa = 4.14LDD348 pKa = 4.16VLSDD352 pKa = 3.39IVLEE356 pKa = 4.09YY357 pKa = 10.97SNSGEE362 pKa = 4.28TIPWAQNYY370 pKa = 8.96YY371 pKa = 8.72PASVVTNDD379 pKa = 4.37FMPAAQDD386 pKa = 3.51FFLDD390 pKa = 3.99EE391 pKa = 5.0NISGEE396 pKa = 4.51DD397 pKa = 4.04FIQLFDD403 pKa = 4.24EE404 pKa = 5.0AWQNAVKK411 pKa = 10.58
MM1 pKa = 7.52KK2 pKa = 10.5LKK4 pKa = 10.68SFWVALLLGLVLVLSACSGGDD25 pKa = 3.35SDD27 pKa = 6.36DD28 pKa = 4.62EE29 pKa = 4.39SAEE32 pKa = 4.34SNGEE36 pKa = 4.12KK37 pKa = 8.98ITIFQTKK44 pKa = 9.59VEE46 pKa = 4.09ISEE49 pKa = 4.1QMEE52 pKa = 4.22EE53 pKa = 3.6LAEE56 pKa = 4.25IYY58 pKa = 10.45EE59 pKa = 4.33EE60 pKa = 4.16EE61 pKa = 4.18TGVEE65 pKa = 4.15VEE67 pKa = 4.2VWGTTGDD74 pKa = 4.73DD75 pKa = 3.85YY76 pKa = 11.41FQQLQTRR83 pKa = 11.84LNSNQGPSILSLQDD97 pKa = 3.06YY98 pKa = 10.44KK99 pKa = 11.02KK100 pKa = 10.99AEE102 pKa = 4.23NIKK105 pKa = 10.72SYY107 pKa = 10.82LHH109 pKa = 6.77DD110 pKa = 4.2LSGEE114 pKa = 4.03EE115 pKa = 4.68FIDD118 pKa = 4.47NIAPNMAVEE127 pKa = 4.09IDD129 pKa = 4.01GEE131 pKa = 4.88VVGIPYY137 pKa = 10.19GVEE140 pKa = 3.97GFGLVYY146 pKa = 10.9NKK148 pKa = 10.72DD149 pKa = 3.62LVSEE153 pKa = 4.45EE154 pKa = 5.26DD155 pKa = 3.32IQDD158 pKa = 3.21YY159 pKa = 11.3DD160 pKa = 3.96SFVNTLEE167 pKa = 4.03RR168 pKa = 11.84LSNEE172 pKa = 3.89GVDD175 pKa = 3.96PLTLSSEE182 pKa = 4.29AYY184 pKa = 10.13FLIGHH189 pKa = 6.36MSNYY193 pKa = 9.28PFSLQEE199 pKa = 3.81DD200 pKa = 3.75HH201 pKa = 7.47FEE203 pKa = 4.69YY204 pKa = 10.08IDD206 pKa = 3.53QLSEE210 pKa = 4.48GEE212 pKa = 4.29VTMTEE217 pKa = 3.9TPEE220 pKa = 3.74FQEE223 pKa = 3.94FGKK226 pKa = 10.34FLEE229 pKa = 5.09AIRR232 pKa = 11.84EE233 pKa = 4.02YY234 pKa = 11.09SPNPMDD240 pKa = 3.41YY241 pKa = 10.29TYY243 pKa = 10.81DD244 pKa = 3.43EE245 pKa = 4.22QMGDD249 pKa = 3.29FATGKK254 pKa = 7.99TAMVHH259 pKa = 4.85QGNWAMGMLADD270 pKa = 3.89YY271 pKa = 10.66DD272 pKa = 4.14YY273 pKa = 11.39DD274 pKa = 4.31FEE276 pKa = 5.21VGMMPFPLAGNDD288 pKa = 3.36KK289 pKa = 10.58LAVGVGANWAVNGTKK304 pKa = 10.42DD305 pKa = 3.32EE306 pKa = 4.54AEE308 pKa = 3.91IEE310 pKa = 4.18AAVDD314 pKa = 3.75FLEE317 pKa = 4.2WLHH320 pKa = 6.0TSEE323 pKa = 3.78TGQRR327 pKa = 11.84FVVEE331 pKa = 4.03EE332 pKa = 4.28FGFVPAMTNIEE343 pKa = 4.28ANDD346 pKa = 4.14LDD348 pKa = 4.16VLSDD352 pKa = 3.39IVLEE356 pKa = 4.09YY357 pKa = 10.97SNSGEE362 pKa = 4.28TIPWAQNYY370 pKa = 8.96YY371 pKa = 8.72PASVVTNDD379 pKa = 4.37FMPAAQDD386 pKa = 3.51FFLDD390 pKa = 3.99EE391 pKa = 5.0NISGEE396 pKa = 4.51DD397 pKa = 4.04FIQLFDD403 pKa = 4.24EE404 pKa = 5.0AWQNAVKK411 pKa = 10.58
Molecular weight: 46.1 kDa
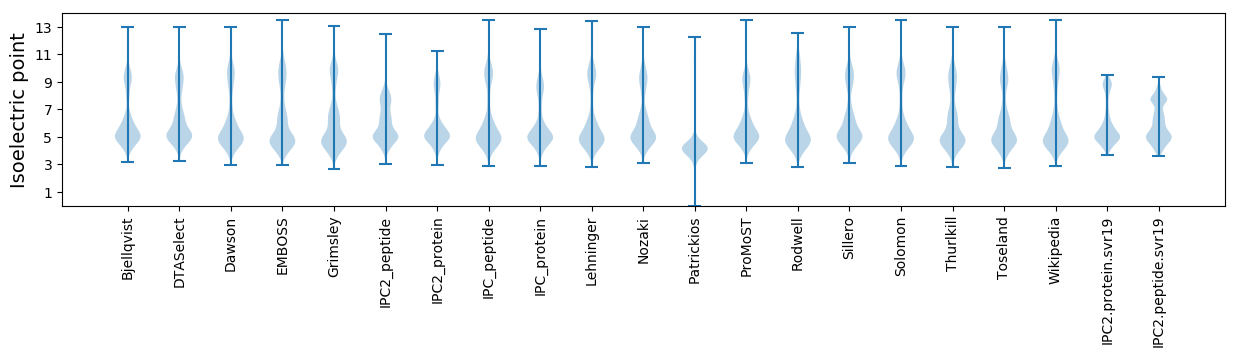
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9LJK3|A0A1H9LJK3_9BACI Uncharacterized protein OS=Piscibacillus halophilus OX=571933 GN=SAMN05216362_14817 PE=4 SV=1
MM1 pKa = 7.42SFLGPHH7 pKa = 6.7LGFGHH12 pKa = 5.77FHH14 pKa = 7.07GFRR17 pKa = 11.84PHH19 pKa = 5.45PRR21 pKa = 11.84FRR23 pKa = 11.84RR24 pKa = 11.84FHH26 pKa = 6.54GFGHH30 pKa = 6.7HH31 pKa = 6.48PRR33 pKa = 11.84FGRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GFGRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GFGHH48 pKa = 6.13HH49 pKa = 6.87HH50 pKa = 5.88GFKK53 pKa = 10.69PFFF56 pKa = 4.41
MM1 pKa = 7.42SFLGPHH7 pKa = 6.7LGFGHH12 pKa = 5.77FHH14 pKa = 7.07GFRR17 pKa = 11.84PHH19 pKa = 5.45PRR21 pKa = 11.84FRR23 pKa = 11.84RR24 pKa = 11.84FHH26 pKa = 6.54GFGHH30 pKa = 6.7HH31 pKa = 6.48PRR33 pKa = 11.84FGRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GFGRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GFGHH48 pKa = 6.13HH49 pKa = 6.87HH50 pKa = 5.88GFKK53 pKa = 10.69PFFF56 pKa = 4.41
Molecular weight: 6.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
874420 |
26 |
3022 |
278.5 |
31.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.262 ± 0.041 | 0.583 ± 0.013 |
5.709 ± 0.043 | 8.061 ± 0.064 |
4.5 ± 0.042 | 6.736 ± 0.044 |
2.292 ± 0.021 | 7.863 ± 0.045 |
6.332 ± 0.048 | 9.625 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.765 ± 0.022 | 4.603 ± 0.033 |
3.564 ± 0.025 | 4.176 ± 0.032 |
3.929 ± 0.033 | 5.874 ± 0.032 |
5.24 ± 0.027 | 7.199 ± 0.039 |
1.029 ± 0.017 | 3.658 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
