
Human immunodeficiency virus type 2 subtype B (isolate UC1) (HIV-2)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Lentivirus; Human immunodeficiency virus 2; HIV-2 subtype B
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
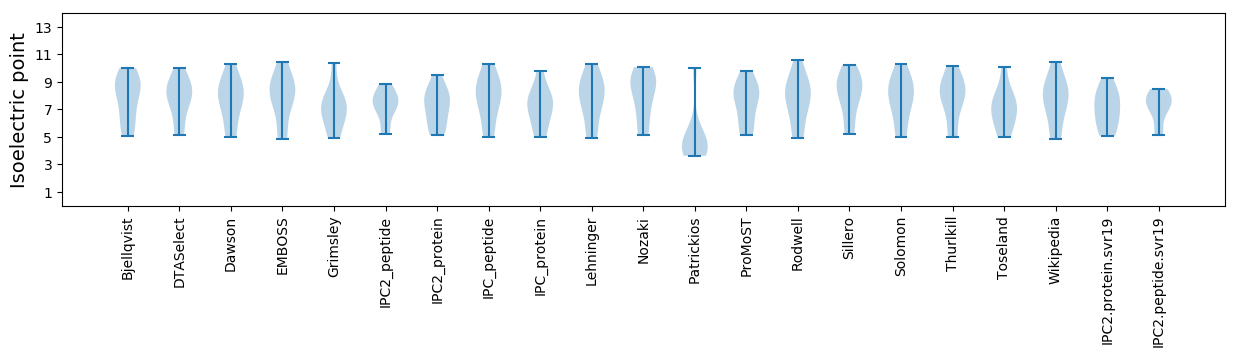
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|Q76638|ENV_HV2UC Envelope glycoprotein gp160 OS=Human immunodeficiency virus type 2 subtype B (isolate UC1) OX=388822 GN=env PE=3 SV=1
MM1 pKa = 7.65AEE3 pKa = 4.61AAPEE7 pKa = 4.04TPPEE11 pKa = 4.14NEE13 pKa = 4.1SPQRR17 pKa = 11.84EE18 pKa = 4.11PWEE21 pKa = 3.98EE22 pKa = 3.7WVEE25 pKa = 4.07DD26 pKa = 3.5VMEE29 pKa = 4.85EE30 pKa = 3.9IKK32 pKa = 10.8QEE34 pKa = 4.05ALRR37 pKa = 11.84HH38 pKa = 5.31FDD40 pKa = 3.54PRR42 pKa = 11.84LLTALGNFIYY52 pKa = 10.37SRR54 pKa = 11.84HH55 pKa = 5.95GDD57 pKa = 3.5TLAGAGEE64 pKa = 4.73LIKK67 pKa = 10.37ILQRR71 pKa = 11.84ALFLHH76 pKa = 6.52FRR78 pKa = 11.84AGCQHH83 pKa = 6.08SRR85 pKa = 11.84IGQPGGGNPLSAIPPSS101 pKa = 3.47
MM1 pKa = 7.65AEE3 pKa = 4.61AAPEE7 pKa = 4.04TPPEE11 pKa = 4.14NEE13 pKa = 4.1SPQRR17 pKa = 11.84EE18 pKa = 4.11PWEE21 pKa = 3.98EE22 pKa = 3.7WVEE25 pKa = 4.07DD26 pKa = 3.5VMEE29 pKa = 4.85EE30 pKa = 3.9IKK32 pKa = 10.8QEE34 pKa = 4.05ALRR37 pKa = 11.84HH38 pKa = 5.31FDD40 pKa = 3.54PRR42 pKa = 11.84LLTALGNFIYY52 pKa = 10.37SRR54 pKa = 11.84HH55 pKa = 5.95GDD57 pKa = 3.5TLAGAGEE64 pKa = 4.73LIKK67 pKa = 10.37ILQRR71 pKa = 11.84ALFLHH76 pKa = 6.52FRR78 pKa = 11.84AGCQHH83 pKa = 6.08SRR85 pKa = 11.84IGQPGGGNPLSAIPPSS101 pKa = 3.47
Molecular weight: 11.23 kDa
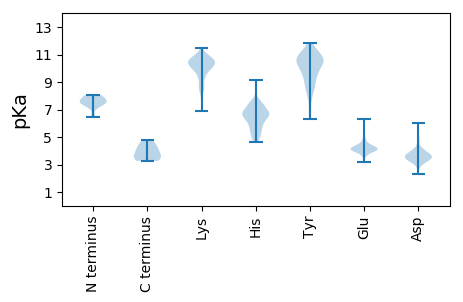
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q76636|VPX_HV2UC Protein Vpx OS=Human immunodeficiency virus type 2 subtype B (isolate UC1) OX=388822 GN=vpx PE=3 SV=1
MM1 pKa = 7.57EE2 pKa = 5.37GEE4 pKa = 4.47KK5 pKa = 10.49NWIVVPTWRR14 pKa = 11.84IPGRR18 pKa = 11.84LEE20 pKa = 3.54KK21 pKa = 9.76WHH23 pKa = 6.86SLVKK27 pKa = 10.2YY28 pKa = 9.76LKK30 pKa = 10.37HH31 pKa = 5.59RR32 pKa = 11.84TKK34 pKa = 10.73EE35 pKa = 4.04LQQVSYY41 pKa = 10.92VPHH44 pKa = 6.83HH45 pKa = 6.05KK46 pKa = 10.78VGWAWWTCSRR56 pKa = 11.84VIFPLKK62 pKa = 10.1EE63 pKa = 3.45EE64 pKa = 4.93AYY66 pKa = 10.96LEE68 pKa = 4.25VQGYY72 pKa = 8.36WNLTPEE78 pKa = 4.22RR79 pKa = 11.84GFLSSYY85 pKa = 10.58AVRR88 pKa = 11.84LTWYY92 pKa = 9.84KK93 pKa = 10.43RR94 pKa = 11.84SFYY97 pKa = 10.77TDD99 pKa = 3.15VTPDD103 pKa = 3.08VADD106 pKa = 3.62QLLHH110 pKa = 6.63GSYY113 pKa = 10.55FSCFTANEE121 pKa = 3.53VRR123 pKa = 11.84RR124 pKa = 11.84AIRR127 pKa = 11.84GEE129 pKa = 4.35KK130 pKa = 9.22ILSYY134 pKa = 10.81CNYY137 pKa = 9.14PSAHH141 pKa = 6.74EE142 pKa = 4.22GQVPSLQFLALRR154 pKa = 11.84VIQEE158 pKa = 4.49GKK160 pKa = 10.22DD161 pKa = 3.59GSQGEE166 pKa = 4.53SATRR170 pKa = 11.84KK171 pKa = 7.55QRR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84NNRR178 pKa = 11.84RR179 pKa = 11.84SIRR182 pKa = 11.84LARR185 pKa = 11.84KK186 pKa = 9.09NNNRR190 pKa = 11.84AQQGSSQPLAPRR202 pKa = 11.84THH204 pKa = 6.54FPGLAEE210 pKa = 4.07VLGILAA216 pKa = 4.81
MM1 pKa = 7.57EE2 pKa = 5.37GEE4 pKa = 4.47KK5 pKa = 10.49NWIVVPTWRR14 pKa = 11.84IPGRR18 pKa = 11.84LEE20 pKa = 3.54KK21 pKa = 9.76WHH23 pKa = 6.86SLVKK27 pKa = 10.2YY28 pKa = 9.76LKK30 pKa = 10.37HH31 pKa = 5.59RR32 pKa = 11.84TKK34 pKa = 10.73EE35 pKa = 4.04LQQVSYY41 pKa = 10.92VPHH44 pKa = 6.83HH45 pKa = 6.05KK46 pKa = 10.78VGWAWWTCSRR56 pKa = 11.84VIFPLKK62 pKa = 10.1EE63 pKa = 3.45EE64 pKa = 4.93AYY66 pKa = 10.96LEE68 pKa = 4.25VQGYY72 pKa = 8.36WNLTPEE78 pKa = 4.22RR79 pKa = 11.84GFLSSYY85 pKa = 10.58AVRR88 pKa = 11.84LTWYY92 pKa = 9.84KK93 pKa = 10.43RR94 pKa = 11.84SFYY97 pKa = 10.77TDD99 pKa = 3.15VTPDD103 pKa = 3.08VADD106 pKa = 3.62QLLHH110 pKa = 6.63GSYY113 pKa = 10.55FSCFTANEE121 pKa = 3.53VRR123 pKa = 11.84RR124 pKa = 11.84AIRR127 pKa = 11.84GEE129 pKa = 4.35KK130 pKa = 9.22ILSYY134 pKa = 10.81CNYY137 pKa = 9.14PSAHH141 pKa = 6.74EE142 pKa = 4.22GQVPSLQFLALRR154 pKa = 11.84VIQEE158 pKa = 4.49GKK160 pKa = 10.22DD161 pKa = 3.59GSQGEE166 pKa = 4.53SATRR170 pKa = 11.84KK171 pKa = 7.55QRR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84NNRR178 pKa = 11.84RR179 pKa = 11.84SIRR182 pKa = 11.84LARR185 pKa = 11.84KK186 pKa = 9.09NNNRR190 pKa = 11.84AQQGSSQPLAPRR202 pKa = 11.84THH204 pKa = 6.54FPGLAEE210 pKa = 4.07VLGILAA216 pKa = 4.81
Molecular weight: 25.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3877 |
97 |
1471 |
387.7 |
43.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.397 ± 0.477 | 2.399 ± 0.35 |
3.998 ± 0.335 | 7.248 ± 0.597 |
2.502 ± 0.239 | 7.274 ± 0.53 |
2.167 ± 0.212 | 5.236 ± 0.668 |
6.345 ± 0.678 | 8.512 ± 0.357 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.218 ± 0.309 | 4.436 ± 0.571 |
6.216 ± 0.721 | 5.726 ± 0.316 |
6.242 ± 0.739 | 5.52 ± 1.211 |
6.319 ± 0.877 | 5.623 ± 0.703 |
2.45 ± 0.266 | 3.173 ± 0.381 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
