
Helicobasidium mompa totivirus 1-17
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus
Average proteome isoelectric point is 8.07
Get precalculated fractions of proteins
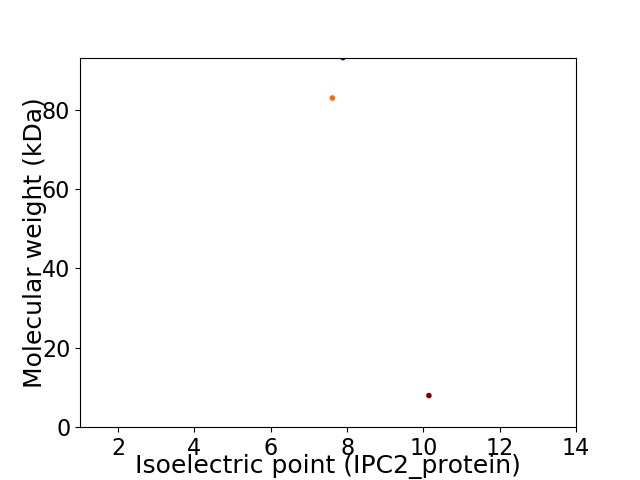
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
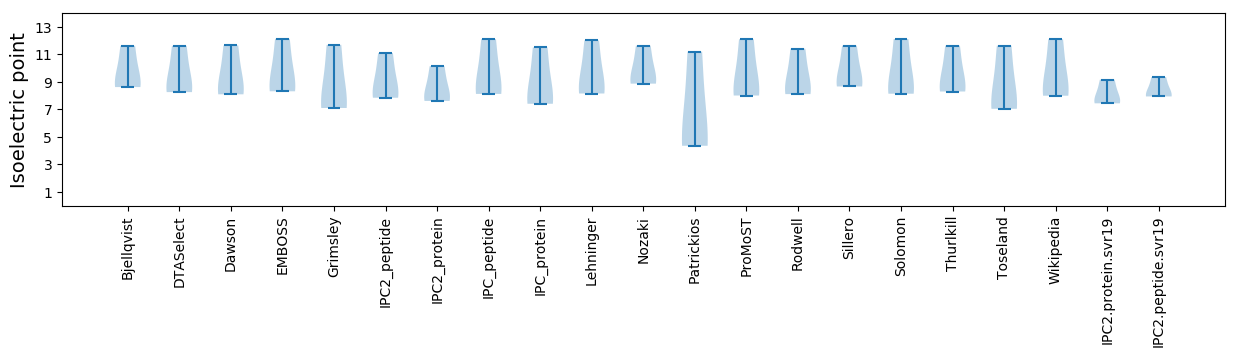
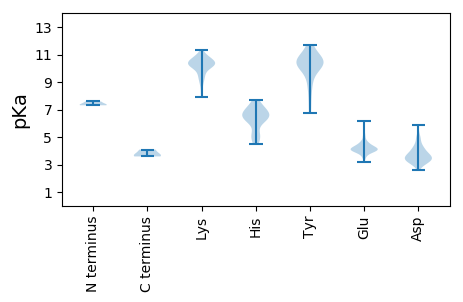
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q76L28|Q76L28_9VIRU Coat protein OS=Helicobasidium mompa totivirus 1-17 OX=196690 GN=CP PE=4 SV=1
MM1 pKa = 7.59SSTSEE6 pKa = 4.25TNQVNYY12 pKa = 10.19LAGVIANPRR21 pKa = 11.84GGQMGQQYY29 pKa = 10.09RR30 pKa = 11.84STVSTITTMAKK41 pKa = 10.01VNSNADD47 pKa = 3.3SRR49 pKa = 11.84SSVVKK54 pKa = 9.09YY55 pKa = 10.26EE56 pKa = 3.22IGRR59 pKa = 11.84RR60 pKa = 11.84HH61 pKa = 6.19DD62 pKa = 3.21ARR64 pKa = 11.84RR65 pKa = 11.84NAFAPYY71 pKa = 9.12ADD73 pKa = 3.43EE74 pKa = 5.31AIRR77 pKa = 11.84VDD79 pKa = 3.59VSYY82 pKa = 8.11DD83 pKa = 3.17TPAIMSEE90 pKa = 4.29TFAGLAKK97 pKa = 10.24KK98 pKa = 10.36YY99 pKa = 10.48SNFSASFEE107 pKa = 4.13RR108 pKa = 11.84SSLAGIVEE116 pKa = 4.46RR117 pKa = 11.84LAKK120 pKa = 10.33VSRR123 pKa = 11.84CKK125 pKa = 10.51LLPRR129 pKa = 11.84CYY131 pKa = 9.85IGGFDD136 pKa = 3.64RR137 pKa = 11.84RR138 pKa = 11.84ARR140 pKa = 11.84DD141 pKa = 3.57PVTGLATHH149 pKa = 6.55FTPVSSLDD157 pKa = 3.22ASVFIPRR164 pKa = 11.84RR165 pKa = 11.84IDD167 pKa = 2.96TMTCPGVFAVLCAAVAGEE185 pKa = 4.62GSQIVTDD192 pKa = 4.87LIEE195 pKa = 4.63INVGNNHH202 pKa = 7.2PIVTTVDD209 pKa = 3.53VAGLPAACVGALRR222 pKa = 11.84VLGANMIAAGQGDD235 pKa = 4.46LFGYY239 pKa = 10.25AVARR243 pKa = 11.84GIHH246 pKa = 4.73QVVSVVGHH254 pKa = 5.13TDD256 pKa = 2.72EE257 pKa = 5.11GGVMRR262 pKa = 11.84DD263 pKa = 3.5LLRR266 pKa = 11.84VDD268 pKa = 4.4GFGAPFGGIHH278 pKa = 6.78FGLPVYY284 pKa = 10.1TGLPHH289 pKa = 7.61IDD291 pKa = 3.45VGNDD295 pKa = 3.19LQLSGWVDD303 pKa = 4.95GIALSTAALVAHH315 pKa = 7.25CDD317 pKa = 3.07HH318 pKa = 6.23GVVYY322 pKa = 10.75NGTWFPTTLSGSAAAPVEE340 pKa = 4.4PGLDD344 pKa = 3.25APAGDD349 pKa = 3.8EE350 pKa = 4.67GYY352 pKa = 10.64VARR355 pKa = 11.84NRR357 pKa = 11.84NQLADD362 pKa = 3.81EE363 pKa = 4.98LSPFSLNYY371 pKa = 10.79ANGLAKK377 pKa = 10.76LFGAGTAGGLTVLHH391 pKa = 6.49LSGRR395 pKa = 11.84AGSLADD401 pKa = 4.11DD402 pKa = 3.79CRR404 pKa = 11.84HH405 pKa = 6.48LNFATANPFFWIEE418 pKa = 4.09STSILPANFLGTQAEE433 pKa = 4.67TEE435 pKa = 4.18GRR437 pKa = 11.84ASYY440 pKa = 10.71GGRR443 pKa = 11.84CSNSSLPLWNQATCVSAVDD462 pKa = 3.92SAVCEE467 pKa = 4.35YY468 pKa = 9.01TVEE471 pKa = 4.13MASPRR476 pKa = 11.84RR477 pKa = 11.84VPFMLHH483 pKa = 5.23WNGNPRR489 pKa = 11.84NGLGNILPRR498 pKa = 11.84QMDD501 pKa = 4.05PMAIIQPGGAEE512 pKa = 3.48ARR514 pKa = 11.84SIRR517 pKa = 11.84EE518 pKa = 3.53RR519 pKa = 11.84MIANNDD525 pKa = 3.37LASYY529 pKa = 10.19LWTRR533 pKa = 11.84GQSPIAAPGEE543 pKa = 4.03FMNTSINVGILVKK556 pKa = 10.49HH557 pKa = 6.77CEE559 pKa = 4.4TTWDD563 pKa = 3.87GAVLSHH569 pKa = 6.75LPSPSEE575 pKa = 4.01FTTGALWLQVGRR587 pKa = 11.84PTGVAVGPLGKK598 pKa = 9.01TNANEE603 pKa = 3.68RR604 pKa = 11.84RR605 pKa = 11.84ARR607 pKa = 11.84TAATNALLRR616 pKa = 11.84ARR618 pKa = 11.84HH619 pKa = 6.12LSRR622 pKa = 11.84GHH624 pKa = 6.07NLVAVNMLPLSFSAPSFGAPRR645 pKa = 11.84VIEE648 pKa = 3.85THH650 pKa = 7.03DD651 pKa = 4.27NINRR655 pKa = 11.84APPGGGSADD664 pKa = 3.3TGVGVDD670 pKa = 3.51RR671 pKa = 11.84VVNAADD677 pKa = 3.34AQGPLRR683 pKa = 11.84PTGRR687 pKa = 11.84TTSNGCPSRR696 pKa = 11.84QGDD699 pKa = 3.59QTWRR703 pKa = 11.84VAVHH707 pKa = 6.47GLSLSPSTLLRR718 pKa = 11.84RR719 pKa = 11.84CLEE722 pKa = 4.31GGCATRR728 pKa = 11.84SVAAACRR735 pKa = 11.84RR736 pKa = 11.84DD737 pKa = 3.52SCTGPRR743 pKa = 11.84NGGGAASGRR752 pKa = 11.84GCRR755 pKa = 11.84TVGPRR760 pKa = 11.84MGPTTFRR767 pKa = 11.84YY768 pKa = 9.68SRR770 pKa = 11.84HH771 pKa = 5.18EE772 pKa = 4.16AEE774 pKa = 6.14AMQQDD779 pKa = 4.05AALQAAGAQQ788 pKa = 3.59
MM1 pKa = 7.59SSTSEE6 pKa = 4.25TNQVNYY12 pKa = 10.19LAGVIANPRR21 pKa = 11.84GGQMGQQYY29 pKa = 10.09RR30 pKa = 11.84STVSTITTMAKK41 pKa = 10.01VNSNADD47 pKa = 3.3SRR49 pKa = 11.84SSVVKK54 pKa = 9.09YY55 pKa = 10.26EE56 pKa = 3.22IGRR59 pKa = 11.84RR60 pKa = 11.84HH61 pKa = 6.19DD62 pKa = 3.21ARR64 pKa = 11.84RR65 pKa = 11.84NAFAPYY71 pKa = 9.12ADD73 pKa = 3.43EE74 pKa = 5.31AIRR77 pKa = 11.84VDD79 pKa = 3.59VSYY82 pKa = 8.11DD83 pKa = 3.17TPAIMSEE90 pKa = 4.29TFAGLAKK97 pKa = 10.24KK98 pKa = 10.36YY99 pKa = 10.48SNFSASFEE107 pKa = 4.13RR108 pKa = 11.84SSLAGIVEE116 pKa = 4.46RR117 pKa = 11.84LAKK120 pKa = 10.33VSRR123 pKa = 11.84CKK125 pKa = 10.51LLPRR129 pKa = 11.84CYY131 pKa = 9.85IGGFDD136 pKa = 3.64RR137 pKa = 11.84RR138 pKa = 11.84ARR140 pKa = 11.84DD141 pKa = 3.57PVTGLATHH149 pKa = 6.55FTPVSSLDD157 pKa = 3.22ASVFIPRR164 pKa = 11.84RR165 pKa = 11.84IDD167 pKa = 2.96TMTCPGVFAVLCAAVAGEE185 pKa = 4.62GSQIVTDD192 pKa = 4.87LIEE195 pKa = 4.63INVGNNHH202 pKa = 7.2PIVTTVDD209 pKa = 3.53VAGLPAACVGALRR222 pKa = 11.84VLGANMIAAGQGDD235 pKa = 4.46LFGYY239 pKa = 10.25AVARR243 pKa = 11.84GIHH246 pKa = 4.73QVVSVVGHH254 pKa = 5.13TDD256 pKa = 2.72EE257 pKa = 5.11GGVMRR262 pKa = 11.84DD263 pKa = 3.5LLRR266 pKa = 11.84VDD268 pKa = 4.4GFGAPFGGIHH278 pKa = 6.78FGLPVYY284 pKa = 10.1TGLPHH289 pKa = 7.61IDD291 pKa = 3.45VGNDD295 pKa = 3.19LQLSGWVDD303 pKa = 4.95GIALSTAALVAHH315 pKa = 7.25CDD317 pKa = 3.07HH318 pKa = 6.23GVVYY322 pKa = 10.75NGTWFPTTLSGSAAAPVEE340 pKa = 4.4PGLDD344 pKa = 3.25APAGDD349 pKa = 3.8EE350 pKa = 4.67GYY352 pKa = 10.64VARR355 pKa = 11.84NRR357 pKa = 11.84NQLADD362 pKa = 3.81EE363 pKa = 4.98LSPFSLNYY371 pKa = 10.79ANGLAKK377 pKa = 10.76LFGAGTAGGLTVLHH391 pKa = 6.49LSGRR395 pKa = 11.84AGSLADD401 pKa = 4.11DD402 pKa = 3.79CRR404 pKa = 11.84HH405 pKa = 6.48LNFATANPFFWIEE418 pKa = 4.09STSILPANFLGTQAEE433 pKa = 4.67TEE435 pKa = 4.18GRR437 pKa = 11.84ASYY440 pKa = 10.71GGRR443 pKa = 11.84CSNSSLPLWNQATCVSAVDD462 pKa = 3.92SAVCEE467 pKa = 4.35YY468 pKa = 9.01TVEE471 pKa = 4.13MASPRR476 pKa = 11.84RR477 pKa = 11.84VPFMLHH483 pKa = 5.23WNGNPRR489 pKa = 11.84NGLGNILPRR498 pKa = 11.84QMDD501 pKa = 4.05PMAIIQPGGAEE512 pKa = 3.48ARR514 pKa = 11.84SIRR517 pKa = 11.84EE518 pKa = 3.53RR519 pKa = 11.84MIANNDD525 pKa = 3.37LASYY529 pKa = 10.19LWTRR533 pKa = 11.84GQSPIAAPGEE543 pKa = 4.03FMNTSINVGILVKK556 pKa = 10.49HH557 pKa = 6.77CEE559 pKa = 4.4TTWDD563 pKa = 3.87GAVLSHH569 pKa = 6.75LPSPSEE575 pKa = 4.01FTTGALWLQVGRR587 pKa = 11.84PTGVAVGPLGKK598 pKa = 9.01TNANEE603 pKa = 3.68RR604 pKa = 11.84RR605 pKa = 11.84ARR607 pKa = 11.84TAATNALLRR616 pKa = 11.84ARR618 pKa = 11.84HH619 pKa = 6.12LSRR622 pKa = 11.84GHH624 pKa = 6.07NLVAVNMLPLSFSAPSFGAPRR645 pKa = 11.84VIEE648 pKa = 3.85THH650 pKa = 7.03DD651 pKa = 4.27NINRR655 pKa = 11.84APPGGGSADD664 pKa = 3.3TGVGVDD670 pKa = 3.51RR671 pKa = 11.84VVNAADD677 pKa = 3.34AQGPLRR683 pKa = 11.84PTGRR687 pKa = 11.84TTSNGCPSRR696 pKa = 11.84QGDD699 pKa = 3.59QTWRR703 pKa = 11.84VAVHH707 pKa = 6.47GLSLSPSTLLRR718 pKa = 11.84RR719 pKa = 11.84CLEE722 pKa = 4.31GGCATRR728 pKa = 11.84SVAAACRR735 pKa = 11.84RR736 pKa = 11.84DD737 pKa = 3.52SCTGPRR743 pKa = 11.84NGGGAASGRR752 pKa = 11.84GCRR755 pKa = 11.84TVGPRR760 pKa = 11.84MGPTTFRR767 pKa = 11.84YY768 pKa = 9.68SRR770 pKa = 11.84HH771 pKa = 5.18EE772 pKa = 4.16AEE774 pKa = 6.14AMQQDD779 pKa = 4.05AALQAAGAQQ788 pKa = 3.59
Molecular weight: 82.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q76L27|Q76L27_9VIRU RNA-directed RNA polymerase OS=Helicobasidium mompa totivirus 1-17 OX=196690 GN=RDRP PE=3 SV=1
MM1 pKa = 7.31SRR3 pKa = 11.84NSRR6 pKa = 11.84ITLGLGPNAQKK17 pKa = 10.74AAAPRR22 pKa = 11.84FSRR25 pKa = 11.84SIAVPVRR32 pKa = 11.84APSLVQAYY40 pKa = 9.09NWLRR44 pKa = 11.84NDD46 pKa = 3.62QVQKK50 pKa = 11.3ALALPEE56 pKa = 4.15IEE58 pKa = 4.32PPARR62 pKa = 11.84LLSPPPQPNHH72 pKa = 4.47TT73 pKa = 4.09
MM1 pKa = 7.31SRR3 pKa = 11.84NSRR6 pKa = 11.84ITLGLGPNAQKK17 pKa = 10.74AAAPRR22 pKa = 11.84FSRR25 pKa = 11.84SIAVPVRR32 pKa = 11.84APSLVQAYY40 pKa = 9.09NWLRR44 pKa = 11.84NDD46 pKa = 3.62QVQKK50 pKa = 11.3ALALPEE56 pKa = 4.15IEE58 pKa = 4.32PPARR62 pKa = 11.84LLSPPPQPNHH72 pKa = 4.47TT73 pKa = 4.09
Molecular weight: 7.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1706 |
73 |
845 |
568.7 |
61.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.489 ± 0.74 | 2.286 ± 0.572 |
4.631 ± 0.816 | 4.162 ± 0.622 |
2.931 ± 0.395 | 8.558 ± 2.035 |
2.462 ± 0.271 | 4.22 ± 0.274 |
2.989 ± 1.215 | 9.379 ± 1.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.638 ± 0.528 | 4.865 ± 0.516 |
5.217 ± 2.454 | 3.048 ± 0.922 |
7.386 ± 0.572 | 7.268 ± 0.34 |
5.686 ± 0.882 | 7.268 ± 0.506 |
1.348 ± 0.134 | 2.169 ± 0.223 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
