
Mesoflavibacter sabulilitoris
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Mesoflavibacter
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
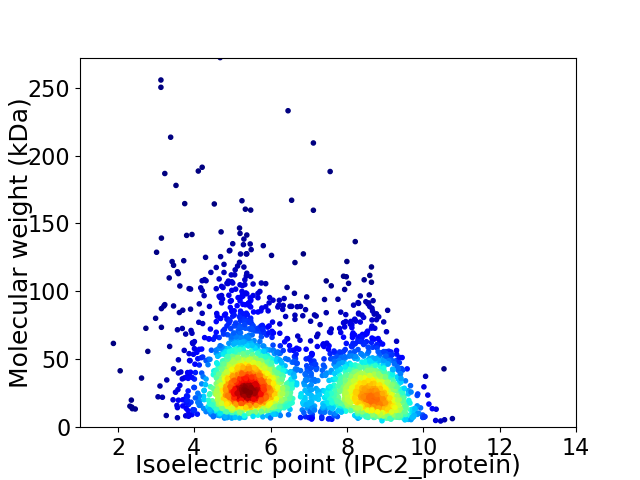
Virtual 2D-PAGE plot for 2809 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T1NIG9|A0A2T1NIG9_9FLAO Cytochrome c oxidase subunit I OS=Mesoflavibacter sabulilitoris OX=1520893 GN=C7H61_04465 PE=3 SV=1
MM1 pKa = 7.29KK2 pKa = 10.26QQLLLTVFILSFFSGLAQNSWKK24 pKa = 10.3KK25 pKa = 10.22ISSNSIEE32 pKa = 4.15NVNLKK37 pKa = 10.54KK38 pKa = 10.47FDD40 pKa = 4.49SNHH43 pKa = 5.87SALFSLDD50 pKa = 2.91IEE52 pKa = 4.27AFKK55 pKa = 11.16AKK57 pKa = 9.96LQSAPLRR64 pKa = 11.84SSSNGQSSTIVSIPDD79 pKa = 3.19EE80 pKa = 4.26TGKK83 pKa = 10.1ILQFRR88 pKa = 11.84VVEE91 pKa = 4.24APVLSKK97 pKa = 10.35EE98 pKa = 5.4LSIQYY103 pKa = 10.14PNIKK107 pKa = 9.18TYY109 pKa = 10.78LGSLVNDD116 pKa = 3.04VTTRR120 pKa = 11.84VRR122 pKa = 11.84FSVTPLGVNVMITRR136 pKa = 11.84ANQEE140 pKa = 3.84LTLIQPVSRR149 pKa = 11.84FSNVEE154 pKa = 3.69HH155 pKa = 6.8IAYY158 pKa = 9.09KK159 pKa = 10.24RR160 pKa = 11.84KK161 pKa = 10.36SINEE165 pKa = 3.69DD166 pKa = 2.99VKK168 pKa = 11.0RR169 pKa = 11.84FEE171 pKa = 4.74CLTDD175 pKa = 3.12EE176 pKa = 4.96TKK178 pKa = 10.41IRR180 pKa = 11.84DD181 pKa = 3.67QNIGQSRR188 pKa = 11.84MMDD191 pKa = 3.33ANDD194 pKa = 3.24QLLRR198 pKa = 11.84DD199 pKa = 3.69YY200 pKa = 10.8RR201 pKa = 11.84IAVSTNGEE209 pKa = 4.13YY210 pKa = 9.48TQFWDD215 pKa = 5.16DD216 pKa = 3.35GNAANGDD223 pKa = 3.85AQQDD227 pKa = 3.27ALAQVVSTLNRR238 pKa = 11.84VNEE241 pKa = 4.21VYY243 pKa = 10.27EE244 pKa = 3.96VDD246 pKa = 3.36MAITFTLVTGTEE258 pKa = 4.29IIFPDD263 pKa = 4.64PATDD267 pKa = 4.21PYY269 pKa = 10.35TGSFNSQLQNEE280 pKa = 4.56LTSTVGEE287 pKa = 3.92ANYY290 pKa = 10.66DD291 pKa = 3.44IGHH294 pKa = 7.04LFAYY298 pKa = 9.97GGNNGNAGCIGCVCEE313 pKa = 5.24DD314 pKa = 3.54NQKK317 pKa = 10.64GSGFSSHH324 pKa = 6.62SFLDD328 pKa = 3.87NDD330 pKa = 3.71GGAYY334 pKa = 9.62MSDD337 pKa = 3.93FFDD340 pKa = 3.54IDD342 pKa = 4.05YY343 pKa = 10.48VPHH346 pKa = 7.08EE347 pKa = 5.17IGHH350 pKa = 5.26QMGANHH356 pKa = 6.59TFAFNTEE363 pKa = 4.08GTGVNSEE370 pKa = 4.47PGSGTTIMGYY380 pKa = 10.88AGITGANDD388 pKa = 3.35VQDD391 pKa = 4.18HH392 pKa = 6.27SDD394 pKa = 3.29AYY396 pKa = 10.31FHH398 pKa = 5.96YY399 pKa = 10.64HH400 pKa = 7.14SINQILTNVASAPNNCATTTVITNNPPVANAGLDD434 pKa = 3.54YY435 pKa = 10.24TIPNGTAFVLKK446 pKa = 10.47GAANDD451 pKa = 3.93ADD453 pKa = 3.98SGDD456 pKa = 3.56VLTYY460 pKa = 9.03TWEE463 pKa = 4.31QIDD466 pKa = 3.93SGSTTSTSFGPAHH479 pKa = 6.79TGPVWRR485 pKa = 11.84SRR487 pKa = 11.84PPSTNPDD494 pKa = 2.74RR495 pKa = 11.84YY496 pKa = 8.66MPIIEE501 pKa = 4.05RR502 pKa = 11.84VIAGEE507 pKa = 3.93LTEE510 pKa = 4.29TNPVEE515 pKa = 4.26TFDD518 pKa = 3.56NSSWEE523 pKa = 4.28TVSTIGRR530 pKa = 11.84TLNFALTVRR539 pKa = 11.84DD540 pKa = 3.54RR541 pKa = 11.84SDD543 pKa = 3.1AGGVGQTPQSDD554 pKa = 4.06FDD556 pKa = 4.15TMTVTVDD563 pKa = 3.61GSSGPFAVTSQTSAVTWDD581 pKa = 3.26AGSTEE586 pKa = 4.41TITWDD591 pKa = 3.38VAGTNTGAVNTPTVNIKK608 pKa = 10.73LSTDD612 pKa = 2.95GGFTYY617 pKa = 9.62PYY619 pKa = 10.79VLATDD624 pKa = 3.84VPNDD628 pKa = 3.64GSHH631 pKa = 7.76DD632 pKa = 3.21ITVPVTGGDD641 pKa = 3.31TATARR646 pKa = 11.84VMVEE650 pKa = 3.58GNNNIFYY657 pKa = 10.49AINSTNFNIQEE668 pKa = 4.17SEE670 pKa = 4.28FVITANNTPVDD681 pKa = 3.75VCSPNDD687 pKa = 3.34AVFNFTYY694 pKa = 9.58NTFLGFSGTTTFSTSGLPAGTTATITPTTATADD727 pKa = 3.66GTAVTVTVTGIGGLSIGNYY746 pKa = 9.37PFTLTGTSGSITSSANVEE764 pKa = 4.34FNVNDD769 pKa = 4.07TNFSTINTLTPALGVTDD786 pKa = 4.03VAPSNAVFTWDD797 pKa = 4.28LDD799 pKa = 4.15PNATSYY805 pKa = 11.15EE806 pKa = 3.96LDD808 pKa = 3.16IATDD812 pKa = 3.41SGFTSIIASQTLTANNYY829 pKa = 7.86TSTSLATQTMYY840 pKa = 8.47WWRR843 pKa = 11.84VRR845 pKa = 11.84AVNDD849 pKa = 4.13CGSGAYY855 pKa = 7.73ATSNFEE861 pKa = 4.25TANIACNTYY870 pKa = 9.53TSNDD874 pKa = 3.48TPIAIPDD881 pKa = 3.86SNTTGISSIITVTDD895 pKa = 3.21VSSIIDD901 pKa = 3.48VNVTINITHH910 pKa = 6.64SYY912 pKa = 10.72VSDD915 pKa = 3.5LTVSLVSPAGTEE927 pKa = 3.37IDD929 pKa = 3.84LSFEE933 pKa = 4.24NGGSGMNYY941 pKa = 8.81TDD943 pKa = 3.87TVFDD947 pKa = 5.28DD948 pKa = 4.96DD949 pKa = 4.4AANPIASGTAPFTGVYY965 pKa = 9.79QPTEE969 pKa = 3.78ALSILNSEE977 pKa = 4.79FSSGDD982 pKa = 2.88WTLKK986 pKa = 10.91VIDD989 pKa = 4.62SYY991 pKa = 12.18GFDD994 pKa = 3.49TGDD997 pKa = 3.32LVNWSIEE1004 pKa = 4.07ICGSPQNDD1012 pKa = 3.45TDD1014 pKa = 4.37GDD1016 pKa = 4.48GIPDD1020 pKa = 3.92GSDD1023 pKa = 2.84NCVNTPNFDD1032 pKa = 3.54QADD1035 pKa = 3.5VDD1037 pKa = 3.91GDD1039 pKa = 4.47GIGDD1043 pKa = 3.75VCDD1046 pKa = 5.41DD1047 pKa = 5.68DD1048 pKa = 5.06IDD1050 pKa = 4.42NDD1052 pKa = 4.98GILNTSDD1059 pKa = 2.88NCIYY1063 pKa = 10.31TPNADD1068 pKa = 3.55QADD1071 pKa = 3.78ADD1073 pKa = 4.1GDD1075 pKa = 4.53GIGDD1079 pKa = 3.73VCDD1082 pKa = 3.99VEE1084 pKa = 4.6CTVYY1088 pKa = 10.76SATDD1092 pKa = 3.64LPITIASTGADD1103 pKa = 3.13ANYY1106 pKa = 9.82TSVINILEE1114 pKa = 4.48DD1115 pKa = 3.79APIADD1120 pKa = 3.96VNVTISINHH1129 pKa = 5.72TWNSDD1134 pKa = 3.16LDD1136 pKa = 3.52ISLISPAGTIIEE1148 pKa = 4.45LTTGNGGSSDD1158 pKa = 3.72NYY1160 pKa = 9.17TNTVFDD1166 pKa = 3.76QDD1168 pKa = 3.33ATASITTGSAPFTGVFMPEE1187 pKa = 3.82GDD1189 pKa = 3.81LSLLNGEE1196 pKa = 4.32MSAGNWTLSVDD1207 pKa = 3.7DD1208 pKa = 4.54TYY1210 pKa = 11.85GPEE1213 pKa = 4.32DD1214 pKa = 3.88GGTILSFDD1222 pKa = 4.53LEE1224 pKa = 4.45LCLFSALSIGNDD1236 pKa = 3.1VLNNTEE1242 pKa = 3.94FSVYY1246 pKa = 10.05PNPNNGEE1253 pKa = 3.91FTIKK1257 pKa = 9.7MKK1259 pKa = 9.82NTVFKK1264 pKa = 10.2EE1265 pKa = 3.84DD1266 pKa = 3.47VEE1268 pKa = 4.53IYY1270 pKa = 10.67VYY1272 pKa = 10.22DD1273 pKa = 3.22IRR1275 pKa = 11.84GRR1277 pKa = 11.84KK1278 pKa = 8.76VFYY1281 pKa = 10.72QSFKK1285 pKa = 10.47NSNNFIQQIRR1295 pKa = 11.84LNDD1298 pKa = 3.76VEE1300 pKa = 4.52SGMYY1304 pKa = 9.49ILNVTDD1310 pKa = 4.5GEE1312 pKa = 4.36NTKK1315 pKa = 9.01TKK1317 pKa = 10.77KK1318 pKa = 10.78LIIDD1322 pKa = 3.93
MM1 pKa = 7.29KK2 pKa = 10.26QQLLLTVFILSFFSGLAQNSWKK24 pKa = 10.3KK25 pKa = 10.22ISSNSIEE32 pKa = 4.15NVNLKK37 pKa = 10.54KK38 pKa = 10.47FDD40 pKa = 4.49SNHH43 pKa = 5.87SALFSLDD50 pKa = 2.91IEE52 pKa = 4.27AFKK55 pKa = 11.16AKK57 pKa = 9.96LQSAPLRR64 pKa = 11.84SSSNGQSSTIVSIPDD79 pKa = 3.19EE80 pKa = 4.26TGKK83 pKa = 10.1ILQFRR88 pKa = 11.84VVEE91 pKa = 4.24APVLSKK97 pKa = 10.35EE98 pKa = 5.4LSIQYY103 pKa = 10.14PNIKK107 pKa = 9.18TYY109 pKa = 10.78LGSLVNDD116 pKa = 3.04VTTRR120 pKa = 11.84VRR122 pKa = 11.84FSVTPLGVNVMITRR136 pKa = 11.84ANQEE140 pKa = 3.84LTLIQPVSRR149 pKa = 11.84FSNVEE154 pKa = 3.69HH155 pKa = 6.8IAYY158 pKa = 9.09KK159 pKa = 10.24RR160 pKa = 11.84KK161 pKa = 10.36SINEE165 pKa = 3.69DD166 pKa = 2.99VKK168 pKa = 11.0RR169 pKa = 11.84FEE171 pKa = 4.74CLTDD175 pKa = 3.12EE176 pKa = 4.96TKK178 pKa = 10.41IRR180 pKa = 11.84DD181 pKa = 3.67QNIGQSRR188 pKa = 11.84MMDD191 pKa = 3.33ANDD194 pKa = 3.24QLLRR198 pKa = 11.84DD199 pKa = 3.69YY200 pKa = 10.8RR201 pKa = 11.84IAVSTNGEE209 pKa = 4.13YY210 pKa = 9.48TQFWDD215 pKa = 5.16DD216 pKa = 3.35GNAANGDD223 pKa = 3.85AQQDD227 pKa = 3.27ALAQVVSTLNRR238 pKa = 11.84VNEE241 pKa = 4.21VYY243 pKa = 10.27EE244 pKa = 3.96VDD246 pKa = 3.36MAITFTLVTGTEE258 pKa = 4.29IIFPDD263 pKa = 4.64PATDD267 pKa = 4.21PYY269 pKa = 10.35TGSFNSQLQNEE280 pKa = 4.56LTSTVGEE287 pKa = 3.92ANYY290 pKa = 10.66DD291 pKa = 3.44IGHH294 pKa = 7.04LFAYY298 pKa = 9.97GGNNGNAGCIGCVCEE313 pKa = 5.24DD314 pKa = 3.54NQKK317 pKa = 10.64GSGFSSHH324 pKa = 6.62SFLDD328 pKa = 3.87NDD330 pKa = 3.71GGAYY334 pKa = 9.62MSDD337 pKa = 3.93FFDD340 pKa = 3.54IDD342 pKa = 4.05YY343 pKa = 10.48VPHH346 pKa = 7.08EE347 pKa = 5.17IGHH350 pKa = 5.26QMGANHH356 pKa = 6.59TFAFNTEE363 pKa = 4.08GTGVNSEE370 pKa = 4.47PGSGTTIMGYY380 pKa = 10.88AGITGANDD388 pKa = 3.35VQDD391 pKa = 4.18HH392 pKa = 6.27SDD394 pKa = 3.29AYY396 pKa = 10.31FHH398 pKa = 5.96YY399 pKa = 10.64HH400 pKa = 7.14SINQILTNVASAPNNCATTTVITNNPPVANAGLDD434 pKa = 3.54YY435 pKa = 10.24TIPNGTAFVLKK446 pKa = 10.47GAANDD451 pKa = 3.93ADD453 pKa = 3.98SGDD456 pKa = 3.56VLTYY460 pKa = 9.03TWEE463 pKa = 4.31QIDD466 pKa = 3.93SGSTTSTSFGPAHH479 pKa = 6.79TGPVWRR485 pKa = 11.84SRR487 pKa = 11.84PPSTNPDD494 pKa = 2.74RR495 pKa = 11.84YY496 pKa = 8.66MPIIEE501 pKa = 4.05RR502 pKa = 11.84VIAGEE507 pKa = 3.93LTEE510 pKa = 4.29TNPVEE515 pKa = 4.26TFDD518 pKa = 3.56NSSWEE523 pKa = 4.28TVSTIGRR530 pKa = 11.84TLNFALTVRR539 pKa = 11.84DD540 pKa = 3.54RR541 pKa = 11.84SDD543 pKa = 3.1AGGVGQTPQSDD554 pKa = 4.06FDD556 pKa = 4.15TMTVTVDD563 pKa = 3.61GSSGPFAVTSQTSAVTWDD581 pKa = 3.26AGSTEE586 pKa = 4.41TITWDD591 pKa = 3.38VAGTNTGAVNTPTVNIKK608 pKa = 10.73LSTDD612 pKa = 2.95GGFTYY617 pKa = 9.62PYY619 pKa = 10.79VLATDD624 pKa = 3.84VPNDD628 pKa = 3.64GSHH631 pKa = 7.76DD632 pKa = 3.21ITVPVTGGDD641 pKa = 3.31TATARR646 pKa = 11.84VMVEE650 pKa = 3.58GNNNIFYY657 pKa = 10.49AINSTNFNIQEE668 pKa = 4.17SEE670 pKa = 4.28FVITANNTPVDD681 pKa = 3.75VCSPNDD687 pKa = 3.34AVFNFTYY694 pKa = 9.58NTFLGFSGTTTFSTSGLPAGTTATITPTTATADD727 pKa = 3.66GTAVTVTVTGIGGLSIGNYY746 pKa = 9.37PFTLTGTSGSITSSANVEE764 pKa = 4.34FNVNDD769 pKa = 4.07TNFSTINTLTPALGVTDD786 pKa = 4.03VAPSNAVFTWDD797 pKa = 4.28LDD799 pKa = 4.15PNATSYY805 pKa = 11.15EE806 pKa = 3.96LDD808 pKa = 3.16IATDD812 pKa = 3.41SGFTSIIASQTLTANNYY829 pKa = 7.86TSTSLATQTMYY840 pKa = 8.47WWRR843 pKa = 11.84VRR845 pKa = 11.84AVNDD849 pKa = 4.13CGSGAYY855 pKa = 7.73ATSNFEE861 pKa = 4.25TANIACNTYY870 pKa = 9.53TSNDD874 pKa = 3.48TPIAIPDD881 pKa = 3.86SNTTGISSIITVTDD895 pKa = 3.21VSSIIDD901 pKa = 3.48VNVTINITHH910 pKa = 6.64SYY912 pKa = 10.72VSDD915 pKa = 3.5LTVSLVSPAGTEE927 pKa = 3.37IDD929 pKa = 3.84LSFEE933 pKa = 4.24NGGSGMNYY941 pKa = 8.81TDD943 pKa = 3.87TVFDD947 pKa = 5.28DD948 pKa = 4.96DD949 pKa = 4.4AANPIASGTAPFTGVYY965 pKa = 9.79QPTEE969 pKa = 3.78ALSILNSEE977 pKa = 4.79FSSGDD982 pKa = 2.88WTLKK986 pKa = 10.91VIDD989 pKa = 4.62SYY991 pKa = 12.18GFDD994 pKa = 3.49TGDD997 pKa = 3.32LVNWSIEE1004 pKa = 4.07ICGSPQNDD1012 pKa = 3.45TDD1014 pKa = 4.37GDD1016 pKa = 4.48GIPDD1020 pKa = 3.92GSDD1023 pKa = 2.84NCVNTPNFDD1032 pKa = 3.54QADD1035 pKa = 3.5VDD1037 pKa = 3.91GDD1039 pKa = 4.47GIGDD1043 pKa = 3.75VCDD1046 pKa = 5.41DD1047 pKa = 5.68DD1048 pKa = 5.06IDD1050 pKa = 4.42NDD1052 pKa = 4.98GILNTSDD1059 pKa = 2.88NCIYY1063 pKa = 10.31TPNADD1068 pKa = 3.55QADD1071 pKa = 3.78ADD1073 pKa = 4.1GDD1075 pKa = 4.53GIGDD1079 pKa = 3.73VCDD1082 pKa = 3.99VEE1084 pKa = 4.6CTVYY1088 pKa = 10.76SATDD1092 pKa = 3.64LPITIASTGADD1103 pKa = 3.13ANYY1106 pKa = 9.82TSVINILEE1114 pKa = 4.48DD1115 pKa = 3.79APIADD1120 pKa = 3.96VNVTISINHH1129 pKa = 5.72TWNSDD1134 pKa = 3.16LDD1136 pKa = 3.52ISLISPAGTIIEE1148 pKa = 4.45LTTGNGGSSDD1158 pKa = 3.72NYY1160 pKa = 9.17TNTVFDD1166 pKa = 3.76QDD1168 pKa = 3.33ATASITTGSAPFTGVFMPEE1187 pKa = 3.82GDD1189 pKa = 3.81LSLLNGEE1196 pKa = 4.32MSAGNWTLSVDD1207 pKa = 3.7DD1208 pKa = 4.54TYY1210 pKa = 11.85GPEE1213 pKa = 4.32DD1214 pKa = 3.88GGTILSFDD1222 pKa = 4.53LEE1224 pKa = 4.45LCLFSALSIGNDD1236 pKa = 3.1VLNNTEE1242 pKa = 3.94FSVYY1246 pKa = 10.05PNPNNGEE1253 pKa = 3.91FTIKK1257 pKa = 9.7MKK1259 pKa = 9.82NTVFKK1264 pKa = 10.2EE1265 pKa = 3.84DD1266 pKa = 3.47VEE1268 pKa = 4.53IYY1270 pKa = 10.67VYY1272 pKa = 10.22DD1273 pKa = 3.22IRR1275 pKa = 11.84GRR1277 pKa = 11.84KK1278 pKa = 8.76VFYY1281 pKa = 10.72QSFKK1285 pKa = 10.47NSNNFIQQIRR1295 pKa = 11.84LNDD1298 pKa = 3.76VEE1300 pKa = 4.52SGMYY1304 pKa = 9.49ILNVTDD1310 pKa = 4.5GEE1312 pKa = 4.36NTKK1315 pKa = 9.01TKK1317 pKa = 10.77KK1318 pKa = 10.78LIIDD1322 pKa = 3.93
Molecular weight: 141.19 kDa
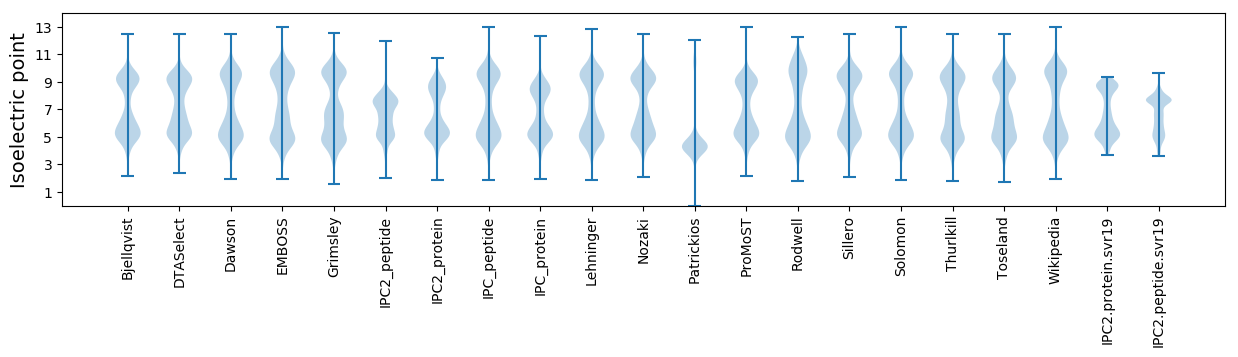
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T1NKN2|A0A2T1NKN2_9FLAO MAM protein (Fragment) OS=Mesoflavibacter sabulilitoris OX=1520893 GN=C7H61_02525 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
940475 |
38 |
2410 |
334.8 |
37.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.127 ± 0.044 | 0.744 ± 0.018 |
5.888 ± 0.04 | 6.447 ± 0.044 |
5.296 ± 0.037 | 5.97 ± 0.05 |
1.686 ± 0.022 | 8.05 ± 0.045 |
8.12 ± 0.083 | 9.217 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.991 ± 0.023 | 6.842 ± 0.053 |
3.238 ± 0.03 | 3.589 ± 0.03 |
3.04 ± 0.029 | 6.289 ± 0.037 |
6.202 ± 0.058 | 6.174 ± 0.033 |
0.95 ± 0.016 | 4.14 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
