
Rhodococcus phage REQ3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
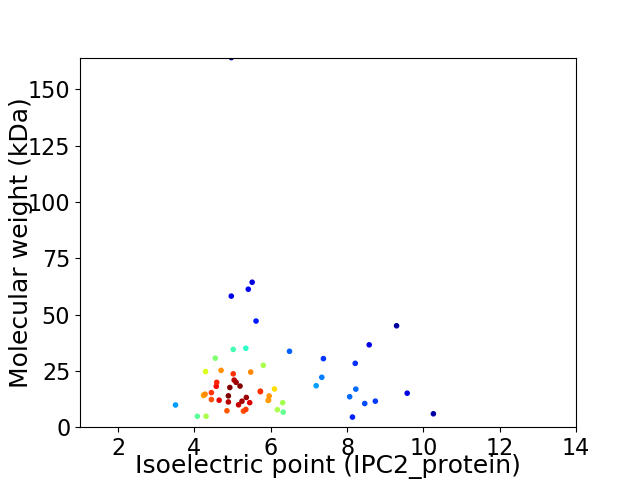
Virtual 2D-PAGE plot for 60 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G9FH64|G9FH64_9CAUD Uncharacterized protein OS=Rhodococcus phage REQ3 OX=1109714 PE=4 SV=1
MM1 pKa = 6.38QTFIEE6 pKa = 4.33RR7 pKa = 11.84GEE9 pKa = 4.38VIALDD14 pKa = 4.65CSHH17 pKa = 6.92QDD19 pKa = 2.97TILFYY24 pKa = 10.84DD25 pKa = 4.34PSYY28 pKa = 11.4GPYY31 pKa = 8.99EE32 pKa = 4.1KK33 pKa = 10.44CVEE36 pKa = 4.23CEE38 pKa = 4.15VILDD42 pKa = 4.19EE43 pKa = 5.4EE44 pKa = 4.9DD45 pKa = 3.9ALADD49 pKa = 3.57NDD51 pKa = 4.05LTDD54 pKa = 3.89EE55 pKa = 4.49EE56 pKa = 5.04VVADD60 pKa = 3.88AGVWVAIWLSALAWTAVVGLGLVWWAVTAA89 pKa = 4.27
MM1 pKa = 6.38QTFIEE6 pKa = 4.33RR7 pKa = 11.84GEE9 pKa = 4.38VIALDD14 pKa = 4.65CSHH17 pKa = 6.92QDD19 pKa = 2.97TILFYY24 pKa = 10.84DD25 pKa = 4.34PSYY28 pKa = 11.4GPYY31 pKa = 8.99EE32 pKa = 4.1KK33 pKa = 10.44CVEE36 pKa = 4.23CEE38 pKa = 4.15VILDD42 pKa = 4.19EE43 pKa = 5.4EE44 pKa = 4.9DD45 pKa = 3.9ALADD49 pKa = 3.57NDD51 pKa = 4.05LTDD54 pKa = 3.89EE55 pKa = 4.49EE56 pKa = 5.04VVADD60 pKa = 3.88AGVWVAIWLSALAWTAVVGLGLVWWAVTAA89 pKa = 4.27
Molecular weight: 9.87 kDa
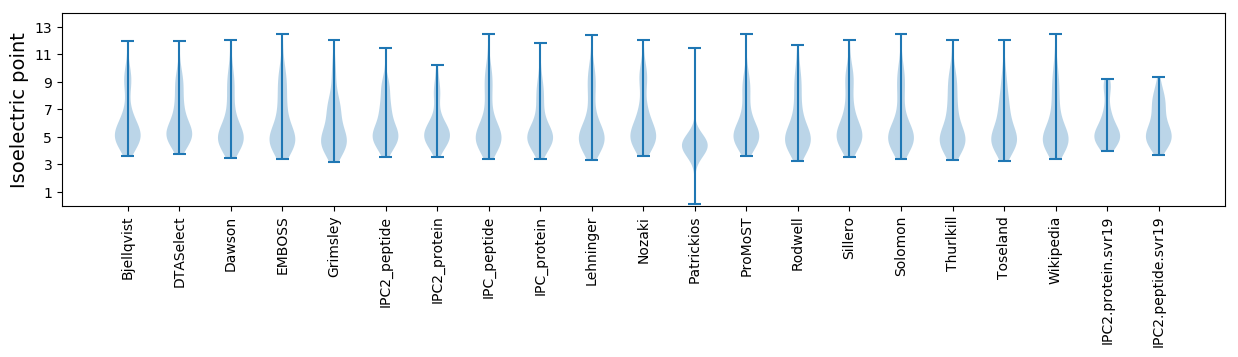
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G9FH49|G9FH49_9CAUD Uncharacterized protein OS=Rhodococcus phage REQ3 OX=1109714 PE=4 SV=1
MM1 pKa = 7.77PRR3 pKa = 11.84PSLPVGAHH11 pKa = 4.91GRR13 pKa = 11.84ISRR16 pKa = 11.84TKK18 pKa = 10.64LPDD21 pKa = 3.5GRR23 pKa = 11.84WRR25 pKa = 11.84AACRR29 pKa = 11.84FRR31 pKa = 11.84DD32 pKa = 3.44ADD34 pKa = 3.55GVTRR38 pKa = 11.84QVVRR42 pKa = 11.84YY43 pKa = 6.86TPPTVDD49 pKa = 3.46RR50 pKa = 11.84DD51 pKa = 3.71KK52 pKa = 11.25TGAAAEE58 pKa = 4.13RR59 pKa = 11.84ALVDD63 pKa = 3.61ALKK66 pKa = 10.83GRR68 pKa = 11.84STTGDD73 pKa = 3.49LSADD77 pKa = 3.49SRR79 pKa = 11.84VSEE82 pKa = 3.76LWMAYY87 pKa = 9.71RR88 pKa = 11.84AQLEE92 pKa = 4.41EE93 pKa = 4.51KK94 pKa = 10.34NRR96 pKa = 11.84SQSTLQDD103 pKa = 3.73YY104 pKa = 11.32DD105 pKa = 4.06RR106 pKa = 11.84MAAKK110 pKa = 10.21ILDD113 pKa = 3.68GLGNLRR119 pKa = 11.84VRR121 pKa = 11.84EE122 pKa = 4.21ATTQRR127 pKa = 11.84LDD129 pKa = 3.31TFVRR133 pKa = 11.84EE134 pKa = 3.55IATRR138 pKa = 11.84QGAGTGKK145 pKa = 10.0KK146 pKa = 10.25AKK148 pKa = 9.47TILSGMFRR156 pKa = 11.84IAVRR160 pKa = 11.84YY161 pKa = 8.89GAVQANPVRR170 pKa = 11.84EE171 pKa = 4.25VTDD174 pKa = 4.0LGAGRR179 pKa = 11.84KK180 pKa = 8.82KK181 pKa = 10.09RR182 pKa = 11.84AKK184 pKa = 10.85SMDD187 pKa = 3.56RR188 pKa = 11.84EE189 pKa = 4.25LLVQLLADD197 pKa = 3.82VRR199 pKa = 11.84GSEE202 pKa = 4.35APCPVVLSEE211 pKa = 4.04AQIKK215 pKa = 10.48RR216 pKa = 11.84GVKK219 pKa = 6.35TTSKK223 pKa = 10.25AGQVPSVAQFCQAADD238 pKa = 3.71LADD241 pKa = 5.16LIVMFAATGARR252 pKa = 11.84IGEE255 pKa = 4.07VLGIRR260 pKa = 11.84WEE262 pKa = 4.19DD263 pKa = 2.95VDD265 pKa = 4.3LKK267 pKa = 11.06KK268 pKa = 10.31RR269 pKa = 11.84TVAIAGKK276 pKa = 8.74VIRR279 pKa = 11.84VKK281 pKa = 11.01GDD283 pKa = 3.11GLVRR287 pKa = 11.84EE288 pKa = 5.08DD289 pKa = 3.47STKK292 pKa = 10.23TEE294 pKa = 3.74SGLRR298 pKa = 11.84QLPLPGFAVEE308 pKa = 4.12MLEE311 pKa = 4.21KK312 pKa = 10.53RR313 pKa = 11.84LVDD316 pKa = 3.22RR317 pKa = 11.84TGPMVFPSKK326 pKa = 10.74VGTLRR331 pKa = 11.84DD332 pKa = 3.52PDD334 pKa = 3.81TVQRR338 pKa = 11.84QWRR341 pKa = 11.84QVRR344 pKa = 11.84AALDD348 pKa = 3.99LEE350 pKa = 4.58WVTTHH355 pKa = 6.35TFRR358 pKa = 11.84KK359 pKa = 7.48TVATILDD366 pKa = 4.19DD367 pKa = 4.23EE368 pKa = 4.9GLTARR373 pKa = 11.84QAADD377 pKa = 3.45HH378 pKa = 6.79LGHH381 pKa = 6.68AQVSMTQDD389 pKa = 2.98VYY391 pKa = 11.15LGRR394 pKa = 11.84GRR396 pKa = 11.84THH398 pKa = 6.46SAAAAALDD406 pKa = 3.57AAVAKK411 pKa = 10.48RR412 pKa = 3.45
MM1 pKa = 7.77PRR3 pKa = 11.84PSLPVGAHH11 pKa = 4.91GRR13 pKa = 11.84ISRR16 pKa = 11.84TKK18 pKa = 10.64LPDD21 pKa = 3.5GRR23 pKa = 11.84WRR25 pKa = 11.84AACRR29 pKa = 11.84FRR31 pKa = 11.84DD32 pKa = 3.44ADD34 pKa = 3.55GVTRR38 pKa = 11.84QVVRR42 pKa = 11.84YY43 pKa = 6.86TPPTVDD49 pKa = 3.46RR50 pKa = 11.84DD51 pKa = 3.71KK52 pKa = 11.25TGAAAEE58 pKa = 4.13RR59 pKa = 11.84ALVDD63 pKa = 3.61ALKK66 pKa = 10.83GRR68 pKa = 11.84STTGDD73 pKa = 3.49LSADD77 pKa = 3.49SRR79 pKa = 11.84VSEE82 pKa = 3.76LWMAYY87 pKa = 9.71RR88 pKa = 11.84AQLEE92 pKa = 4.41EE93 pKa = 4.51KK94 pKa = 10.34NRR96 pKa = 11.84SQSTLQDD103 pKa = 3.73YY104 pKa = 11.32DD105 pKa = 4.06RR106 pKa = 11.84MAAKK110 pKa = 10.21ILDD113 pKa = 3.68GLGNLRR119 pKa = 11.84VRR121 pKa = 11.84EE122 pKa = 4.21ATTQRR127 pKa = 11.84LDD129 pKa = 3.31TFVRR133 pKa = 11.84EE134 pKa = 3.55IATRR138 pKa = 11.84QGAGTGKK145 pKa = 10.0KK146 pKa = 10.25AKK148 pKa = 9.47TILSGMFRR156 pKa = 11.84IAVRR160 pKa = 11.84YY161 pKa = 8.89GAVQANPVRR170 pKa = 11.84EE171 pKa = 4.25VTDD174 pKa = 4.0LGAGRR179 pKa = 11.84KK180 pKa = 8.82KK181 pKa = 10.09RR182 pKa = 11.84AKK184 pKa = 10.85SMDD187 pKa = 3.56RR188 pKa = 11.84EE189 pKa = 4.25LLVQLLADD197 pKa = 3.82VRR199 pKa = 11.84GSEE202 pKa = 4.35APCPVVLSEE211 pKa = 4.04AQIKK215 pKa = 10.48RR216 pKa = 11.84GVKK219 pKa = 6.35TTSKK223 pKa = 10.25AGQVPSVAQFCQAADD238 pKa = 3.71LADD241 pKa = 5.16LIVMFAATGARR252 pKa = 11.84IGEE255 pKa = 4.07VLGIRR260 pKa = 11.84WEE262 pKa = 4.19DD263 pKa = 2.95VDD265 pKa = 4.3LKK267 pKa = 11.06KK268 pKa = 10.31RR269 pKa = 11.84TVAIAGKK276 pKa = 8.74VIRR279 pKa = 11.84VKK281 pKa = 11.01GDD283 pKa = 3.11GLVRR287 pKa = 11.84EE288 pKa = 5.08DD289 pKa = 3.47STKK292 pKa = 10.23TEE294 pKa = 3.74SGLRR298 pKa = 11.84QLPLPGFAVEE308 pKa = 4.12MLEE311 pKa = 4.21KK312 pKa = 10.53RR313 pKa = 11.84LVDD316 pKa = 3.22RR317 pKa = 11.84TGPMVFPSKK326 pKa = 10.74VGTLRR331 pKa = 11.84DD332 pKa = 3.52PDD334 pKa = 3.81TVQRR338 pKa = 11.84QWRR341 pKa = 11.84QVRR344 pKa = 11.84AALDD348 pKa = 3.99LEE350 pKa = 4.58WVTTHH355 pKa = 6.35TFRR358 pKa = 11.84KK359 pKa = 7.48TVATILDD366 pKa = 4.19DD367 pKa = 4.23EE368 pKa = 4.9GLTARR373 pKa = 11.84QAADD377 pKa = 3.45HH378 pKa = 6.79LGHH381 pKa = 6.68AQVSMTQDD389 pKa = 2.98VYY391 pKa = 11.15LGRR394 pKa = 11.84GRR396 pKa = 11.84THH398 pKa = 6.46SAAAAALDD406 pKa = 3.57AAVAKK411 pKa = 10.48RR412 pKa = 3.45
Molecular weight: 45.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12346 |
40 |
1585 |
205.8 |
22.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.765 ± 0.488 | 0.923 ± 0.205 |
6.707 ± 0.276 | 6.14 ± 0.356 |
2.576 ± 0.178 | 8.57 ± 0.562 |
1.96 ± 0.212 | 4.674 ± 0.245 |
3.532 ± 0.321 | 7.614 ± 0.241 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.236 ± 0.137 | 3.005 ± 0.191 |
5.435 ± 0.337 | 3.515 ± 0.231 |
7.079 ± 0.478 | 5.613 ± 0.251 |
7.031 ± 0.33 | 6.699 ± 0.327 |
1.717 ± 0.15 | 2.211 ± 0.179 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
