
Nonlabens spongiae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Nonlabens
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
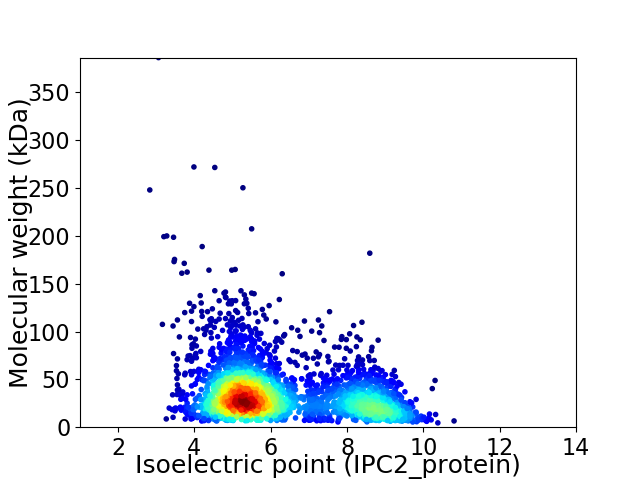
Virtual 2D-PAGE plot for 2989 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
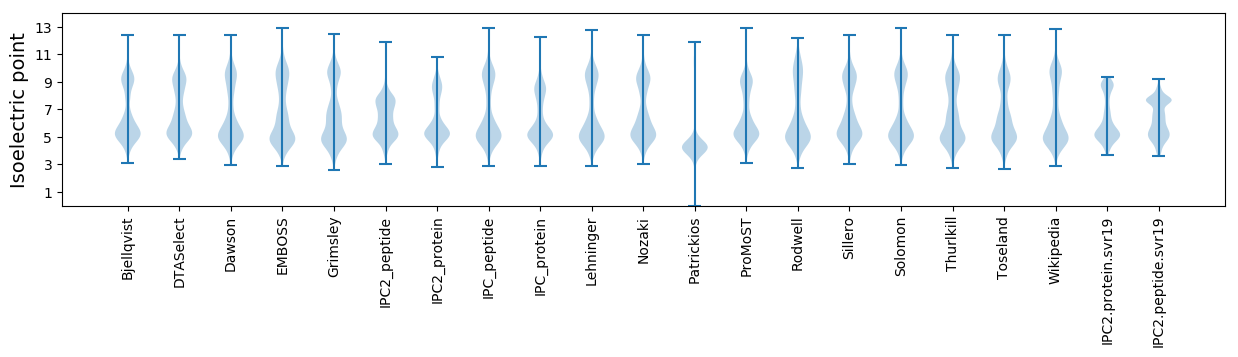
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W6MPD0|A0A1W6MPD0_9FLAO Uncharacterized protein OS=Nonlabens spongiae OX=331648 GN=BST97_11475 PE=4 SV=1
MM1 pKa = 7.25MNKK4 pKa = 9.81FYY6 pKa = 9.79ITLITVSVFTAQSWAQVPSNYY27 pKa = 9.46YY28 pKa = 10.71DD29 pKa = 3.31SAAGLTGYY37 pKa = 10.26ALKK40 pKa = 10.84SEE42 pKa = 4.28LANIISANYY51 pKa = 8.44NAQSYY56 pKa = 11.48DD57 pKa = 3.7DD58 pKa = 5.65LRR60 pKa = 11.84DD61 pKa = 3.62LYY63 pKa = 11.19AISDD67 pKa = 3.48NDD69 pKa = 3.45AYY71 pKa = 11.3YY72 pKa = 11.09DD73 pKa = 3.62NGQQTTTILDD83 pKa = 4.26LYY85 pKa = 10.98SEE87 pKa = 4.72NPNGADD93 pKa = 3.65PYY95 pKa = 10.53TFSATNTNDD104 pKa = 2.96RR105 pKa = 11.84CGNYY109 pKa = 9.88SGEE112 pKa = 3.96GDD114 pKa = 3.58CWNRR118 pKa = 11.84EE119 pKa = 4.08HH120 pKa = 7.55IFPQGFFNQLEE131 pKa = 4.28PMRR134 pKa = 11.84SDD136 pKa = 2.84AHH138 pKa = 6.83HH139 pKa = 6.94VIPTDD144 pKa = 3.37GFVNGGRR151 pKa = 11.84SNLPFGEE158 pKa = 4.06VDD160 pKa = 3.6LSGSGIRR167 pKa = 11.84TYY169 pKa = 11.31QNGSRR174 pKa = 11.84KK175 pKa = 9.98GPSATPGYY183 pKa = 8.62TGDD186 pKa = 3.36VFEE189 pKa = 6.24PIDD192 pKa = 3.88EE193 pKa = 4.55FKK195 pKa = 11.47GDD197 pKa = 3.59IARR200 pKa = 11.84MLLYY204 pKa = 10.32FATRR208 pKa = 11.84YY209 pKa = 8.99EE210 pKa = 4.97DD211 pKa = 3.47RR212 pKa = 11.84FNDD215 pKa = 4.13NRR217 pKa = 11.84WDD219 pKa = 3.91SPNATNDD226 pKa = 3.47PRR228 pKa = 11.84DD229 pKa = 3.5GSQDD233 pKa = 2.94QYY235 pKa = 11.45YY236 pKa = 8.05EE237 pKa = 3.45QWYY240 pKa = 9.99IDD242 pKa = 5.18LLLSWHH248 pKa = 6.37AQDD251 pKa = 4.37PVSQRR256 pKa = 11.84EE257 pKa = 3.42IDD259 pKa = 3.9RR260 pKa = 11.84NNDD263 pKa = 2.35IYY265 pKa = 11.59NFQNNANPYY274 pKa = 8.63IDD276 pKa = 4.42NPQFVDD282 pKa = 4.37MIWNSSQAPSGSIFATLTDD301 pKa = 3.72SYY303 pKa = 11.8NDD305 pKa = 3.7VNSNGYY311 pKa = 9.58DD312 pKa = 3.11AGDD315 pKa = 4.32EE316 pKa = 4.04INYY319 pKa = 10.45DD320 pKa = 3.55YY321 pKa = 11.06TIEE324 pKa = 4.11NLGNTTLYY332 pKa = 10.71NVTVTASRR340 pKa = 11.84GNFANTVSPIASIAPGQVINNPFGNLQVVLITQDD374 pKa = 2.9VDD376 pKa = 3.69PNGACDD382 pKa = 5.05VINQLQVTADD392 pKa = 4.2FSANEE397 pKa = 4.06NTGGLSIASDD407 pKa = 4.08DD408 pKa = 3.94PDD410 pKa = 3.52NFQDD414 pKa = 4.15VDD416 pKa = 4.49SNNDD420 pKa = 3.56NLPDD424 pKa = 4.82DD425 pKa = 3.95PTISNVCGGSTGTVSEE441 pKa = 4.68LFISEE446 pKa = 4.47YY447 pKa = 10.45IEE449 pKa = 4.63GSGSNKK455 pKa = 10.16AIEE458 pKa = 4.14IANFTGSQVNLSGYY472 pKa = 9.7SIEE475 pKa = 4.54RR476 pKa = 11.84NANGGSTWSGTISLSGTLDD495 pKa = 3.19NGEE498 pKa = 4.4VYY500 pKa = 10.89VLARR504 pKa = 11.84GNADD508 pKa = 3.34QAILDD513 pKa = 3.88EE514 pKa = 4.59ADD516 pKa = 3.61RR517 pKa = 11.84LIGNGNALDD526 pKa = 4.16FNGNDD531 pKa = 3.41PVGLFRR537 pKa = 11.84NGNLIDD543 pKa = 3.48IVGVFNSGSTDD554 pKa = 3.65FAKK557 pKa = 10.8DD558 pKa = 3.32VVLVRR563 pKa = 11.84KK564 pKa = 9.41PDD566 pKa = 3.52AVVPNLDD573 pKa = 3.77FNLTRR578 pKa = 11.84DD579 pKa = 3.5WNSFGQSNYY588 pKa = 9.46TDD590 pKa = 4.43LGQHH594 pKa = 5.37TVTTASEE601 pKa = 4.06DD602 pKa = 3.52QLIAEE607 pKa = 4.53SFKK610 pKa = 11.2VYY612 pKa = 10.36PNPSKK617 pKa = 11.08DD618 pKa = 3.17GIFYY622 pKa = 10.86FEE624 pKa = 4.28TDD626 pKa = 3.41LEE628 pKa = 4.41EE629 pKa = 5.69IEE631 pKa = 5.66LNVFDD636 pKa = 5.44LSGRR640 pKa = 11.84SISFDD645 pKa = 3.15QTQDD649 pKa = 3.68SIQLEE654 pKa = 4.03QAGIYY659 pKa = 9.92ILTVEE664 pKa = 4.2KK665 pKa = 10.83DD666 pKa = 3.63GNRR669 pKa = 11.84SSLKK673 pKa = 10.47LVLRR677 pKa = 4.67
MM1 pKa = 7.25MNKK4 pKa = 9.81FYY6 pKa = 9.79ITLITVSVFTAQSWAQVPSNYY27 pKa = 9.46YY28 pKa = 10.71DD29 pKa = 3.31SAAGLTGYY37 pKa = 10.26ALKK40 pKa = 10.84SEE42 pKa = 4.28LANIISANYY51 pKa = 8.44NAQSYY56 pKa = 11.48DD57 pKa = 3.7DD58 pKa = 5.65LRR60 pKa = 11.84DD61 pKa = 3.62LYY63 pKa = 11.19AISDD67 pKa = 3.48NDD69 pKa = 3.45AYY71 pKa = 11.3YY72 pKa = 11.09DD73 pKa = 3.62NGQQTTTILDD83 pKa = 4.26LYY85 pKa = 10.98SEE87 pKa = 4.72NPNGADD93 pKa = 3.65PYY95 pKa = 10.53TFSATNTNDD104 pKa = 2.96RR105 pKa = 11.84CGNYY109 pKa = 9.88SGEE112 pKa = 3.96GDD114 pKa = 3.58CWNRR118 pKa = 11.84EE119 pKa = 4.08HH120 pKa = 7.55IFPQGFFNQLEE131 pKa = 4.28PMRR134 pKa = 11.84SDD136 pKa = 2.84AHH138 pKa = 6.83HH139 pKa = 6.94VIPTDD144 pKa = 3.37GFVNGGRR151 pKa = 11.84SNLPFGEE158 pKa = 4.06VDD160 pKa = 3.6LSGSGIRR167 pKa = 11.84TYY169 pKa = 11.31QNGSRR174 pKa = 11.84KK175 pKa = 9.98GPSATPGYY183 pKa = 8.62TGDD186 pKa = 3.36VFEE189 pKa = 6.24PIDD192 pKa = 3.88EE193 pKa = 4.55FKK195 pKa = 11.47GDD197 pKa = 3.59IARR200 pKa = 11.84MLLYY204 pKa = 10.32FATRR208 pKa = 11.84YY209 pKa = 8.99EE210 pKa = 4.97DD211 pKa = 3.47RR212 pKa = 11.84FNDD215 pKa = 4.13NRR217 pKa = 11.84WDD219 pKa = 3.91SPNATNDD226 pKa = 3.47PRR228 pKa = 11.84DD229 pKa = 3.5GSQDD233 pKa = 2.94QYY235 pKa = 11.45YY236 pKa = 8.05EE237 pKa = 3.45QWYY240 pKa = 9.99IDD242 pKa = 5.18LLLSWHH248 pKa = 6.37AQDD251 pKa = 4.37PVSQRR256 pKa = 11.84EE257 pKa = 3.42IDD259 pKa = 3.9RR260 pKa = 11.84NNDD263 pKa = 2.35IYY265 pKa = 11.59NFQNNANPYY274 pKa = 8.63IDD276 pKa = 4.42NPQFVDD282 pKa = 4.37MIWNSSQAPSGSIFATLTDD301 pKa = 3.72SYY303 pKa = 11.8NDD305 pKa = 3.7VNSNGYY311 pKa = 9.58DD312 pKa = 3.11AGDD315 pKa = 4.32EE316 pKa = 4.04INYY319 pKa = 10.45DD320 pKa = 3.55YY321 pKa = 11.06TIEE324 pKa = 4.11NLGNTTLYY332 pKa = 10.71NVTVTASRR340 pKa = 11.84GNFANTVSPIASIAPGQVINNPFGNLQVVLITQDD374 pKa = 2.9VDD376 pKa = 3.69PNGACDD382 pKa = 5.05VINQLQVTADD392 pKa = 4.2FSANEE397 pKa = 4.06NTGGLSIASDD407 pKa = 4.08DD408 pKa = 3.94PDD410 pKa = 3.52NFQDD414 pKa = 4.15VDD416 pKa = 4.49SNNDD420 pKa = 3.56NLPDD424 pKa = 4.82DD425 pKa = 3.95PTISNVCGGSTGTVSEE441 pKa = 4.68LFISEE446 pKa = 4.47YY447 pKa = 10.45IEE449 pKa = 4.63GSGSNKK455 pKa = 10.16AIEE458 pKa = 4.14IANFTGSQVNLSGYY472 pKa = 9.7SIEE475 pKa = 4.54RR476 pKa = 11.84NANGGSTWSGTISLSGTLDD495 pKa = 3.19NGEE498 pKa = 4.4VYY500 pKa = 10.89VLARR504 pKa = 11.84GNADD508 pKa = 3.34QAILDD513 pKa = 3.88EE514 pKa = 4.59ADD516 pKa = 3.61RR517 pKa = 11.84LIGNGNALDD526 pKa = 4.16FNGNDD531 pKa = 3.41PVGLFRR537 pKa = 11.84NGNLIDD543 pKa = 3.48IVGVFNSGSTDD554 pKa = 3.65FAKK557 pKa = 10.8DD558 pKa = 3.32VVLVRR563 pKa = 11.84KK564 pKa = 9.41PDD566 pKa = 3.52AVVPNLDD573 pKa = 3.77FNLTRR578 pKa = 11.84DD579 pKa = 3.5WNSFGQSNYY588 pKa = 9.46TDD590 pKa = 4.43LGQHH594 pKa = 5.37TVTTASEE601 pKa = 4.06DD602 pKa = 3.52QLIAEE607 pKa = 4.53SFKK610 pKa = 11.2VYY612 pKa = 10.36PNPSKK617 pKa = 11.08DD618 pKa = 3.17GIFYY622 pKa = 10.86FEE624 pKa = 4.28TDD626 pKa = 3.41LEE628 pKa = 4.41EE629 pKa = 5.69IEE631 pKa = 5.66LNVFDD636 pKa = 5.44LSGRR640 pKa = 11.84SISFDD645 pKa = 3.15QTQDD649 pKa = 3.68SIQLEE654 pKa = 4.03QAGIYY659 pKa = 9.92ILTVEE664 pKa = 4.2KK665 pKa = 10.83DD666 pKa = 3.63GNRR669 pKa = 11.84SSLKK673 pKa = 10.47LVLRR677 pKa = 4.67
Molecular weight: 74.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6MJN6|A0A1W6MJN6_9FLAO DUF4835 domain-containing protein OS=Nonlabens spongiae OX=331648 GN=BST97_07335 PE=4 SV=1
MM1 pKa = 6.94AQKK4 pKa = 9.31RR5 pKa = 11.84TYY7 pKa = 10.22QPSKK11 pKa = 9.01RR12 pKa = 11.84KK13 pKa = 9.47RR14 pKa = 11.84RR15 pKa = 11.84NKK17 pKa = 9.49HH18 pKa = 3.94GFRR21 pKa = 11.84EE22 pKa = 4.27RR23 pKa = 11.84MASANGRR30 pKa = 11.84KK31 pKa = 9.04VLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 10.09GRR41 pKa = 11.84KK42 pKa = 8.0KK43 pKa = 10.66LSVSTEE49 pKa = 3.69RR50 pKa = 11.84RR51 pKa = 11.84HH52 pKa = 6.27KK53 pKa = 10.62RR54 pKa = 3.14
MM1 pKa = 6.94AQKK4 pKa = 9.31RR5 pKa = 11.84TYY7 pKa = 10.22QPSKK11 pKa = 9.01RR12 pKa = 11.84KK13 pKa = 9.47RR14 pKa = 11.84RR15 pKa = 11.84NKK17 pKa = 9.49HH18 pKa = 3.94GFRR21 pKa = 11.84EE22 pKa = 4.27RR23 pKa = 11.84MASANGRR30 pKa = 11.84KK31 pKa = 9.04VLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 10.09GRR41 pKa = 11.84KK42 pKa = 8.0KK43 pKa = 10.66LSVSTEE49 pKa = 3.69RR50 pKa = 11.84RR51 pKa = 11.84HH52 pKa = 6.27KK53 pKa = 10.62RR54 pKa = 3.14
Molecular weight: 6.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
982028 |
38 |
3707 |
328.5 |
37.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.606 ± 0.045 | 0.744 ± 0.013 |
6.232 ± 0.052 | 6.821 ± 0.047 |
4.977 ± 0.036 | 6.485 ± 0.055 |
1.847 ± 0.025 | 7.349 ± 0.038 |
6.882 ± 0.073 | 9.319 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.33 ± 0.027 | 5.589 ± 0.051 |
3.428 ± 0.024 | 3.715 ± 0.029 |
4.158 ± 0.037 | 6.577 ± 0.042 |
5.565 ± 0.05 | 6.375 ± 0.035 |
1.037 ± 0.017 | 3.966 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
