
Koribacter versatilis (strain Ellin345)
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Acidobacteriia; Acidobacteriales; Acidobacteriaceae; Candidatus Koribacter; Candidatus Koribacter versatilis
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
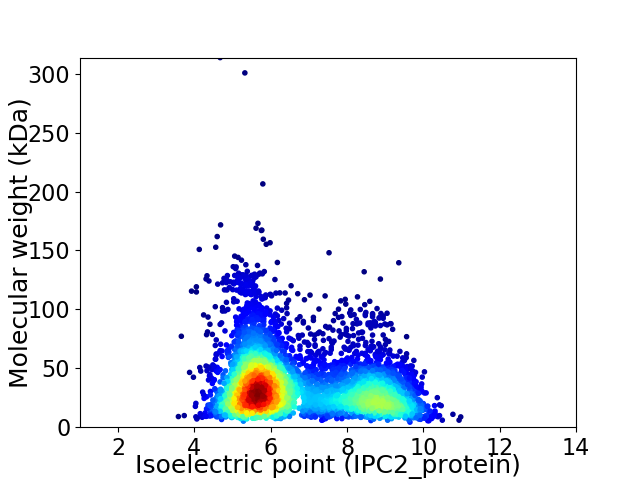
Virtual 2D-PAGE plot for 4771 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1IMF6|Q1IMF6_KORVE Polysulphide reductase NrfD OS=Koribacter versatilis (strain Ellin345) OX=204669 GN=Acid345_2943 PE=3 SV=1
MM1 pKa = 7.23RR2 pKa = 11.84QVAAVSFFLVFTAVFSAYY20 pKa = 10.59SQDD23 pKa = 3.02LTQAKK28 pKa = 7.43RR29 pKa = 11.84TPAASGSTQPNVFLTPKK46 pKa = 9.65QYY48 pKa = 10.08PAGPSGVTSIAKK60 pKa = 10.23GDD62 pKa = 3.95FNNDD66 pKa = 2.2SYY68 pKa = 11.77MDD70 pKa = 3.59VAVTNVSGTITVLLGKK86 pKa = 10.62GDD88 pKa = 3.9GTFQAPVSYY97 pKa = 9.09PALSSPVSIAAADD110 pKa = 3.87LNGDD114 pKa = 3.83GKK116 pKa = 11.32LDD118 pKa = 3.73LAVANSGSGSISVFLGNGDD137 pKa = 3.67GTFQSHH143 pKa = 5.6TDD145 pKa = 3.47VAVGTSVQMLTVADD159 pKa = 4.33FNGDD163 pKa = 3.82GKK165 pKa = 10.11PDD167 pKa = 3.66LAVLVDD173 pKa = 3.62GMVSVLIGKK182 pKa = 9.88GDD184 pKa = 3.42ATFNAIGEE192 pKa = 4.39YY193 pKa = 10.51AKK195 pKa = 10.74PCATYY200 pKa = 10.68LATGDD205 pKa = 3.8FNGDD209 pKa = 3.04GKK211 pKa = 9.5TDD213 pKa = 3.18IVAGRR218 pKa = 11.84QCVLLGNGDD227 pKa = 3.82GTFQPPVGSQKK238 pKa = 10.23IGNTVSTAVGDD249 pKa = 3.75INGDD253 pKa = 3.42GKK255 pKa = 11.3LDD257 pKa = 4.71LIEE260 pKa = 5.21GGIGDD265 pKa = 3.73SDD267 pKa = 3.69GTPRR271 pKa = 11.84ALVVVLLGNGDD282 pKa = 3.81GTFQPPQGFFGYY294 pKa = 10.62GSGVQGLLLADD305 pKa = 3.96VNGDD309 pKa = 3.45SHH311 pKa = 8.07PDD313 pKa = 3.04IVLSSSEE320 pKa = 3.88NVEE323 pKa = 4.25VVNGKK328 pKa = 10.39GDD330 pKa = 3.62GTFEE334 pKa = 4.62PGVLYY339 pKa = 9.87PVGNRR344 pKa = 11.84PVAGGLVLSDD354 pKa = 3.25FTGSGRR360 pKa = 11.84LDD362 pKa = 3.54LAVLTSCANPSICGDD377 pKa = 3.65GAVTLLRR384 pKa = 11.84GKK386 pKa = 11.1GDD388 pKa = 3.53GTYY391 pKa = 10.17VAPASYY397 pKa = 10.83YY398 pKa = 9.91IFEE401 pKa = 4.3GDD403 pKa = 3.29EE404 pKa = 4.0RR405 pKa = 11.84FDD407 pKa = 4.82AVGGWVAVGDD417 pKa = 3.82FDD419 pKa = 6.4GDD421 pKa = 3.63GKK423 pKa = 11.35LDD425 pKa = 3.53VLEE428 pKa = 4.66VFDD431 pKa = 3.82TRR433 pKa = 11.84AFISLGNGDD442 pKa = 3.78GTVQTGQRR450 pKa = 11.84YY451 pKa = 7.86YY452 pKa = 10.65QVAYY456 pKa = 10.24QSDD459 pKa = 4.2GAVVGDD465 pKa = 3.76FNGDD469 pKa = 3.33GKK471 pKa = 11.22LDD473 pKa = 3.54AAILHH478 pKa = 6.2SCDD481 pKa = 3.38VFYY484 pKa = 11.14SDD486 pKa = 5.35PGPGNPPPYY495 pKa = 9.24CVSAGSVGVLLGNGNGTWQRR515 pKa = 11.84WPEE518 pKa = 3.8SLYY521 pKa = 10.94FGVGDD526 pKa = 4.18TPTSIATGDD535 pKa = 3.56FNQDD539 pKa = 2.46GKK541 pKa = 11.51LDD543 pKa = 3.94LVVSDD548 pKa = 4.54GANAYY553 pKa = 9.35ILLGNGDD560 pKa = 3.64GTFPVHH566 pKa = 5.63QAYY569 pKa = 7.26PTGAAADD576 pKa = 3.97YY577 pKa = 10.66LNPFLPNAQSVVVGDD592 pKa = 4.12FNGDD596 pKa = 3.53GAPDD600 pKa = 3.59VAVSNSDD607 pKa = 2.9GGIAVLLGNGDD618 pKa = 3.53GTLRR622 pKa = 11.84APQLFPAIKK631 pKa = 9.93SSQSLAIGDD640 pKa = 4.04LNRR643 pKa = 11.84DD644 pKa = 3.17GKK646 pKa = 11.13LDD648 pKa = 3.41IVASDD653 pKa = 3.79GSGSISIFLGNGDD666 pKa = 3.87GTFQTSKK673 pKa = 10.49VYY675 pKa = 10.57AAVGSQSVTVGDD687 pKa = 4.02FNGDD691 pKa = 3.64GILDD695 pKa = 3.78VASGTGHH702 pKa = 5.69TVSLLLGNGDD712 pKa = 4.23GSLQPPVNYY721 pKa = 9.74IVGLSATGLAAGDD734 pKa = 3.98FNGDD738 pKa = 3.17GALDD742 pKa = 3.91LVTEE746 pKa = 4.75DD747 pKa = 5.85FSILLNRR754 pKa = 11.84QGTQLNVQSSRR765 pKa = 11.84NPSNVGQPVTFTVTSAASLPEE786 pKa = 3.98TGLPSGTITLRR797 pKa = 11.84DD798 pKa = 3.42GSTVLGEE805 pKa = 4.24SGVSGVFDD813 pKa = 3.68VKK815 pKa = 11.16VSGLTAGTHH824 pKa = 5.91QITATYY830 pKa = 10.46SGDD833 pKa = 3.73NNFQPHH839 pKa = 4.7TTAILTEE846 pKa = 4.45HH847 pKa = 6.35VGAPATMISPALGSILTSTTVTFAWKK873 pKa = 9.6AAAGASQYY881 pKa = 11.61SLYY884 pKa = 10.85LGTKK888 pKa = 9.4PGRR891 pKa = 11.84DD892 pKa = 3.28DD893 pKa = 3.79LGYY896 pKa = 11.14VNAHH900 pKa = 6.42SSTSATVKK908 pKa = 10.37NLPSTGSSLYY918 pKa = 8.75VTLFSLVGGVYY929 pKa = 10.29YY930 pKa = 10.86SNSYY934 pKa = 9.58TYY936 pKa = 10.48ILPGTPAKK944 pKa = 10.9AKK946 pKa = 7.3MTSPLPGTMLVGKK959 pKa = 9.61DD960 pKa = 3.36ATFTWSHH967 pKa = 5.34GTGVTYY973 pKa = 10.74YY974 pKa = 10.6SLYY977 pKa = 10.9VGTKK981 pKa = 10.14GYY983 pKa = 7.77GTHH986 pKa = 7.45DD987 pKa = 4.19LDD989 pKa = 5.68FINATTTSASVSNLPADD1006 pKa = 3.85GSTIYY1011 pKa = 10.8VQVNSYY1017 pKa = 10.61IDD1019 pKa = 3.86GAWTSQSYY1027 pKa = 8.68TYY1029 pKa = 10.39ISGSGTPAPATMISPTPGSSISGNSATFTWTSGVGVSEE1067 pKa = 4.32FSLYY1071 pKa = 10.9VGTGGVGSHH1080 pKa = 6.79NIAFIEE1086 pKa = 4.25TGTTSATVTGLPATGATIYY1105 pKa = 10.7VRR1107 pKa = 11.84LNSFVNGAWQWVDD1120 pKa = 2.79YY1121 pKa = 10.71SYY1123 pKa = 11.62RR1124 pKa = 11.84NPP1126 pKa = 3.45
MM1 pKa = 7.23RR2 pKa = 11.84QVAAVSFFLVFTAVFSAYY20 pKa = 10.59SQDD23 pKa = 3.02LTQAKK28 pKa = 7.43RR29 pKa = 11.84TPAASGSTQPNVFLTPKK46 pKa = 9.65QYY48 pKa = 10.08PAGPSGVTSIAKK60 pKa = 10.23GDD62 pKa = 3.95FNNDD66 pKa = 2.2SYY68 pKa = 11.77MDD70 pKa = 3.59VAVTNVSGTITVLLGKK86 pKa = 10.62GDD88 pKa = 3.9GTFQAPVSYY97 pKa = 9.09PALSSPVSIAAADD110 pKa = 3.87LNGDD114 pKa = 3.83GKK116 pKa = 11.32LDD118 pKa = 3.73LAVANSGSGSISVFLGNGDD137 pKa = 3.67GTFQSHH143 pKa = 5.6TDD145 pKa = 3.47VAVGTSVQMLTVADD159 pKa = 4.33FNGDD163 pKa = 3.82GKK165 pKa = 10.11PDD167 pKa = 3.66LAVLVDD173 pKa = 3.62GMVSVLIGKK182 pKa = 9.88GDD184 pKa = 3.42ATFNAIGEE192 pKa = 4.39YY193 pKa = 10.51AKK195 pKa = 10.74PCATYY200 pKa = 10.68LATGDD205 pKa = 3.8FNGDD209 pKa = 3.04GKK211 pKa = 9.5TDD213 pKa = 3.18IVAGRR218 pKa = 11.84QCVLLGNGDD227 pKa = 3.82GTFQPPVGSQKK238 pKa = 10.23IGNTVSTAVGDD249 pKa = 3.75INGDD253 pKa = 3.42GKK255 pKa = 11.3LDD257 pKa = 4.71LIEE260 pKa = 5.21GGIGDD265 pKa = 3.73SDD267 pKa = 3.69GTPRR271 pKa = 11.84ALVVVLLGNGDD282 pKa = 3.81GTFQPPQGFFGYY294 pKa = 10.62GSGVQGLLLADD305 pKa = 3.96VNGDD309 pKa = 3.45SHH311 pKa = 8.07PDD313 pKa = 3.04IVLSSSEE320 pKa = 3.88NVEE323 pKa = 4.25VVNGKK328 pKa = 10.39GDD330 pKa = 3.62GTFEE334 pKa = 4.62PGVLYY339 pKa = 9.87PVGNRR344 pKa = 11.84PVAGGLVLSDD354 pKa = 3.25FTGSGRR360 pKa = 11.84LDD362 pKa = 3.54LAVLTSCANPSICGDD377 pKa = 3.65GAVTLLRR384 pKa = 11.84GKK386 pKa = 11.1GDD388 pKa = 3.53GTYY391 pKa = 10.17VAPASYY397 pKa = 10.83YY398 pKa = 9.91IFEE401 pKa = 4.3GDD403 pKa = 3.29EE404 pKa = 4.0RR405 pKa = 11.84FDD407 pKa = 4.82AVGGWVAVGDD417 pKa = 3.82FDD419 pKa = 6.4GDD421 pKa = 3.63GKK423 pKa = 11.35LDD425 pKa = 3.53VLEE428 pKa = 4.66VFDD431 pKa = 3.82TRR433 pKa = 11.84AFISLGNGDD442 pKa = 3.78GTVQTGQRR450 pKa = 11.84YY451 pKa = 7.86YY452 pKa = 10.65QVAYY456 pKa = 10.24QSDD459 pKa = 4.2GAVVGDD465 pKa = 3.76FNGDD469 pKa = 3.33GKK471 pKa = 11.22LDD473 pKa = 3.54AAILHH478 pKa = 6.2SCDD481 pKa = 3.38VFYY484 pKa = 11.14SDD486 pKa = 5.35PGPGNPPPYY495 pKa = 9.24CVSAGSVGVLLGNGNGTWQRR515 pKa = 11.84WPEE518 pKa = 3.8SLYY521 pKa = 10.94FGVGDD526 pKa = 4.18TPTSIATGDD535 pKa = 3.56FNQDD539 pKa = 2.46GKK541 pKa = 11.51LDD543 pKa = 3.94LVVSDD548 pKa = 4.54GANAYY553 pKa = 9.35ILLGNGDD560 pKa = 3.64GTFPVHH566 pKa = 5.63QAYY569 pKa = 7.26PTGAAADD576 pKa = 3.97YY577 pKa = 10.66LNPFLPNAQSVVVGDD592 pKa = 4.12FNGDD596 pKa = 3.53GAPDD600 pKa = 3.59VAVSNSDD607 pKa = 2.9GGIAVLLGNGDD618 pKa = 3.53GTLRR622 pKa = 11.84APQLFPAIKK631 pKa = 9.93SSQSLAIGDD640 pKa = 4.04LNRR643 pKa = 11.84DD644 pKa = 3.17GKK646 pKa = 11.13LDD648 pKa = 3.41IVASDD653 pKa = 3.79GSGSISIFLGNGDD666 pKa = 3.87GTFQTSKK673 pKa = 10.49VYY675 pKa = 10.57AAVGSQSVTVGDD687 pKa = 4.02FNGDD691 pKa = 3.64GILDD695 pKa = 3.78VASGTGHH702 pKa = 5.69TVSLLLGNGDD712 pKa = 4.23GSLQPPVNYY721 pKa = 9.74IVGLSATGLAAGDD734 pKa = 3.98FNGDD738 pKa = 3.17GALDD742 pKa = 3.91LVTEE746 pKa = 4.75DD747 pKa = 5.85FSILLNRR754 pKa = 11.84QGTQLNVQSSRR765 pKa = 11.84NPSNVGQPVTFTVTSAASLPEE786 pKa = 3.98TGLPSGTITLRR797 pKa = 11.84DD798 pKa = 3.42GSTVLGEE805 pKa = 4.24SGVSGVFDD813 pKa = 3.68VKK815 pKa = 11.16VSGLTAGTHH824 pKa = 5.91QITATYY830 pKa = 10.46SGDD833 pKa = 3.73NNFQPHH839 pKa = 4.7TTAILTEE846 pKa = 4.45HH847 pKa = 6.35VGAPATMISPALGSILTSTTVTFAWKK873 pKa = 9.6AAAGASQYY881 pKa = 11.61SLYY884 pKa = 10.85LGTKK888 pKa = 9.4PGRR891 pKa = 11.84DD892 pKa = 3.28DD893 pKa = 3.79LGYY896 pKa = 11.14VNAHH900 pKa = 6.42SSTSATVKK908 pKa = 10.37NLPSTGSSLYY918 pKa = 8.75VTLFSLVGGVYY929 pKa = 10.29YY930 pKa = 10.86SNSYY934 pKa = 9.58TYY936 pKa = 10.48ILPGTPAKK944 pKa = 10.9AKK946 pKa = 7.3MTSPLPGTMLVGKK959 pKa = 9.61DD960 pKa = 3.36ATFTWSHH967 pKa = 5.34GTGVTYY973 pKa = 10.74YY974 pKa = 10.6SLYY977 pKa = 10.9VGTKK981 pKa = 10.14GYY983 pKa = 7.77GTHH986 pKa = 7.45DD987 pKa = 4.19LDD989 pKa = 5.68FINATTTSASVSNLPADD1006 pKa = 3.85GSTIYY1011 pKa = 10.8VQVNSYY1017 pKa = 10.61IDD1019 pKa = 3.86GAWTSQSYY1027 pKa = 8.68TYY1029 pKa = 10.39ISGSGTPAPATMISPTPGSSISGNSATFTWTSGVGVSEE1067 pKa = 4.32FSLYY1071 pKa = 10.9VGTGGVGSHH1080 pKa = 6.79NIAFIEE1086 pKa = 4.25TGTTSATVTGLPATGATIYY1105 pKa = 10.7VRR1107 pKa = 11.84LNSFVNGAWQWVDD1120 pKa = 2.79YY1121 pKa = 10.71SYY1123 pKa = 11.62RR1124 pKa = 11.84NPP1126 pKa = 3.45
Molecular weight: 114.79 kDa
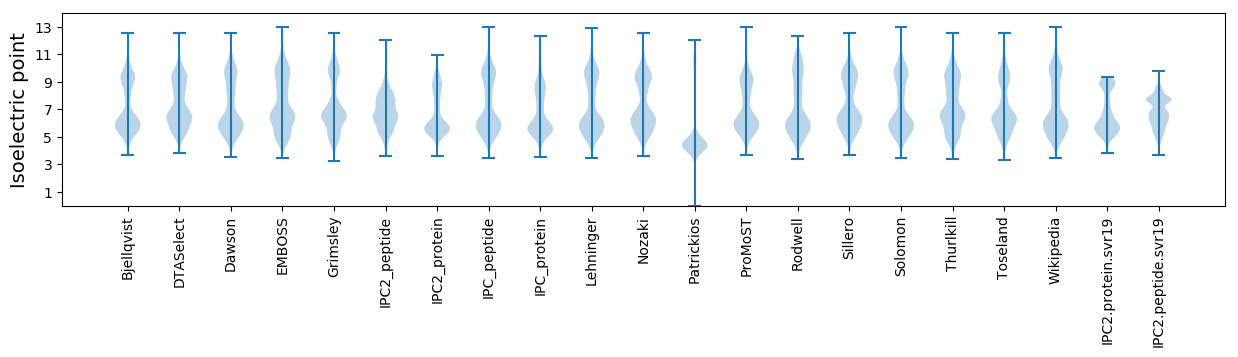
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1IV87|Q1IV87_KORVE Uncharacterized protein OS=Koribacter versatilis (strain Ellin345) OX=204669 GN=Acid345_0208 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.02RR4 pKa = 11.84TFQPNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 8.4THH17 pKa = 5.66GFRR20 pKa = 11.84TRR22 pKa = 11.84MKK24 pKa = 8.81TKK26 pKa = 10.13SGAAVLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.64GRR40 pKa = 11.84KK41 pKa = 8.44RR42 pKa = 11.84VSVSAGYY49 pKa = 10.39RR50 pKa = 11.84DD51 pKa = 3.36
MM1 pKa = 8.0PKK3 pKa = 9.02RR4 pKa = 11.84TFQPNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 8.4THH17 pKa = 5.66GFRR20 pKa = 11.84TRR22 pKa = 11.84MKK24 pKa = 8.81TKK26 pKa = 10.13SGAAVLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.64GRR40 pKa = 11.84KK41 pKa = 8.44RR42 pKa = 11.84VSVSAGYY49 pKa = 10.39RR50 pKa = 11.84DD51 pKa = 3.36
Molecular weight: 5.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1677413 |
37 |
3121 |
351.6 |
38.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.494 ± 0.04 | 0.903 ± 0.012 |
5.257 ± 0.027 | 5.713 ± 0.042 |
4.053 ± 0.023 | 7.895 ± 0.039 |
2.242 ± 0.018 | 5.205 ± 0.027 |
4.473 ± 0.029 | 9.353 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.341 ± 0.018 | 3.542 ± 0.032 |
5.016 ± 0.025 | 3.735 ± 0.022 |
5.969 ± 0.04 | 6.252 ± 0.03 |
5.872 ± 0.038 | 7.52 ± 0.03 |
1.359 ± 0.017 | 2.808 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
