
Torque teno sus virus 1a
Taxonomy: Viruses; Anelloviridae; Iotatorquevirus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
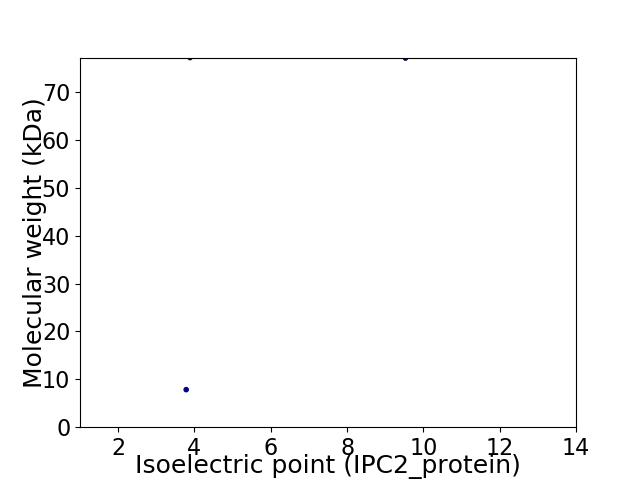
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8TX29|F8TX29_9VIRU ORF2 OS=Torque teno sus virus 1a OX=687386 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.08FLYY4 pKa = 9.97WEE6 pKa = 4.53EE7 pKa = 3.91AWLTAIEE14 pKa = 5.38GYY16 pKa = 10.76HH17 pKa = 6.65NLDD20 pKa = 3.54CRR22 pKa = 11.84CGNWYY27 pKa = 10.07DD28 pKa = 3.46HH29 pKa = 6.1LQRR32 pKa = 11.84LCALDD37 pKa = 3.6QLDD40 pKa = 4.17AVAAAAIEE48 pKa = 4.35RR49 pKa = 11.84EE50 pKa = 4.44DD51 pKa = 4.2GDD53 pKa = 4.0GDD55 pKa = 4.16AATTDD60 pKa = 4.16TITDD64 pKa = 3.84GDD66 pKa = 4.08PGVAGGG72 pKa = 4.02
MM1 pKa = 7.08FLYY4 pKa = 9.97WEE6 pKa = 4.53EE7 pKa = 3.91AWLTAIEE14 pKa = 5.38GYY16 pKa = 10.76HH17 pKa = 6.65NLDD20 pKa = 3.54CRR22 pKa = 11.84CGNWYY27 pKa = 10.07DD28 pKa = 3.46HH29 pKa = 6.1LQRR32 pKa = 11.84LCALDD37 pKa = 3.6QLDD40 pKa = 4.17AVAAAAIEE48 pKa = 4.35RR49 pKa = 11.84EE50 pKa = 4.44DD51 pKa = 4.2GDD53 pKa = 4.0GDD55 pKa = 4.16AATTDD60 pKa = 4.16TITDD64 pKa = 3.84GDD66 pKa = 4.08PGVAGGG72 pKa = 4.02
Molecular weight: 7.85 kDa
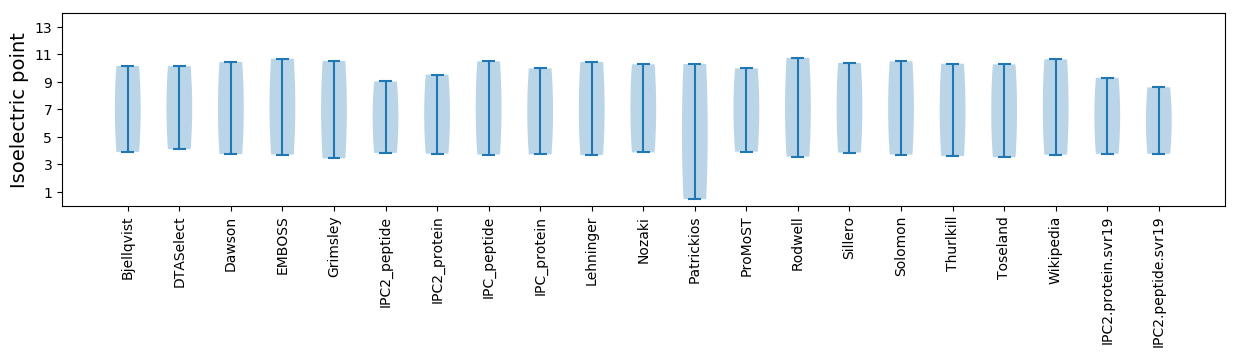
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8TX29|F8TX29_9VIRU ORF2 OS=Torque teno sus virus 1a OX=687386 GN=ORF2 PE=4 SV=1
MM1 pKa = 8.06RR2 pKa = 11.84FRR4 pKa = 11.84PIRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 9.34RR15 pKa = 11.84KK16 pKa = 9.55RR17 pKa = 11.84RR18 pKa = 11.84WGWRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 9.16YY26 pKa = 8.7RR27 pKa = 11.84HH28 pKa = 6.54HH29 pKa = 5.95YY30 pKa = 9.16RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84PWRR36 pKa = 11.84RR37 pKa = 11.84WRR39 pKa = 11.84VRR41 pKa = 11.84RR42 pKa = 11.84WRR44 pKa = 11.84RR45 pKa = 11.84SVFRR49 pKa = 11.84RR50 pKa = 11.84GGRR53 pKa = 11.84RR54 pKa = 11.84ARR56 pKa = 11.84PYY58 pKa = 10.18RR59 pKa = 11.84ISAFNPRR66 pKa = 11.84VMRR69 pKa = 11.84RR70 pKa = 11.84IVIQGWWPLIQCLPGQEE87 pKa = 4.09HH88 pKa = 6.75LRR90 pKa = 11.84YY91 pKa = 9.65RR92 pKa = 11.84PLNWDD97 pKa = 3.3IEE99 pKa = 4.48KK100 pKa = 10.44EE101 pKa = 4.22WVVEE105 pKa = 3.97KK106 pKa = 10.95DD107 pKa = 3.44KK108 pKa = 11.52EE109 pKa = 4.14PDD111 pKa = 2.97KK112 pKa = 10.85MGYY115 pKa = 9.11LVQYY119 pKa = 10.1GGGWGSGHH127 pKa = 6.31ITLEE131 pKa = 4.07DD132 pKa = 4.17LFNEE136 pKa = 4.14HH137 pKa = 5.92QKK139 pKa = 8.42WRR141 pKa = 11.84NMWSKK146 pKa = 11.53SNDD149 pKa = 2.85GMDD152 pKa = 3.68LVRR155 pKa = 11.84YY156 pKa = 7.33FGCKK160 pKa = 8.81IYY162 pKa = 10.43LYY164 pKa = 9.82PLEE167 pKa = 5.55DD168 pKa = 2.88VDD170 pKa = 4.93YY171 pKa = 9.82WFWWDD176 pKa = 3.06TDD178 pKa = 3.77FKK180 pKa = 11.64NNYY183 pKa = 6.99EE184 pKa = 4.07DD185 pKa = 5.33RR186 pKa = 11.84IKK188 pKa = 10.44EE189 pKa = 3.98YY190 pKa = 10.45CQPSVMLMAKK200 pKa = 8.32RR201 pKa = 11.84TKK203 pKa = 10.03IVVARR208 pKa = 11.84KK209 pKa = 8.72RR210 pKa = 11.84AQHH213 pKa = 5.56RR214 pKa = 11.84RR215 pKa = 11.84RR216 pKa = 11.84VKK218 pKa = 10.46KK219 pKa = 10.42IFIPPPSRR227 pKa = 11.84DD228 pKa = 3.04TTQWKK233 pKa = 9.0FQTEE237 pKa = 4.5FCKK240 pKa = 10.7QPLFTWAAGLIEE252 pKa = 4.38LQRR255 pKa = 11.84PFDD258 pKa = 4.05FNGGFRR264 pKa = 11.84NAWWLEE270 pKa = 3.49PRR272 pKa = 11.84NEE274 pKa = 4.27AGEE277 pKa = 4.1MDD279 pKa = 5.56YY280 pKa = 9.86ITLWGRR286 pKa = 11.84VPPQGDD292 pKa = 3.19TDD294 pKa = 3.81MPDD297 pKa = 3.28PNTFKK302 pKa = 8.72TTTQNKK308 pKa = 9.47NYY310 pKa = 9.93SIQPGKK316 pKa = 10.05EE317 pKa = 3.79NNVYY321 pKa = 10.44PIMAYY326 pKa = 9.16IRR328 pKa = 11.84QEE330 pKa = 3.79NQQPVVNWCVCYY342 pKa = 10.89NKK344 pKa = 10.42TIDD347 pKa = 3.15RR348 pKa = 11.84WRR350 pKa = 11.84KK351 pKa = 6.27TQAATLKK358 pKa = 10.18IGNLRR363 pKa = 11.84GLCVRR368 pKa = 11.84EE369 pKa = 4.55LSRR372 pKa = 11.84QEE374 pKa = 3.59MTYY377 pKa = 10.05KK378 pKa = 10.02WKK380 pKa = 10.57QGEE383 pKa = 4.03FSKK386 pKa = 10.55AYY388 pKa = 9.41HH389 pKa = 6.63DD390 pKa = 3.18RR391 pKa = 11.84WGYY394 pKa = 7.7NTYY397 pKa = 10.33FVTIDD402 pKa = 3.37GRR404 pKa = 11.84KK405 pKa = 7.74GTEE408 pKa = 3.89NPTVQIWNFNDD419 pKa = 3.9KK420 pKa = 9.38ITVQSGHH427 pKa = 6.48ILNFAFNIQKK437 pKa = 10.88SNDD440 pKa = 3.41NLKK443 pKa = 11.03DD444 pKa = 3.35SDD446 pKa = 3.89FGKK449 pKa = 10.42IGLPKK454 pKa = 10.24SPHH457 pKa = 6.57DD458 pKa = 4.62LDD460 pKa = 5.33FGHH463 pKa = 7.24KK464 pKa = 9.92NRR466 pKa = 11.84FGPFVVKK473 pKa = 10.77NEE475 pKa = 3.89PVEE478 pKa = 4.23FQLNPPRR485 pKa = 11.84AINVWVRR492 pKa = 11.84YY493 pKa = 10.0KK494 pKa = 10.85FFFQFGGEE502 pKa = 4.11YY503 pKa = 10.14QPPTGIRR510 pKa = 11.84DD511 pKa = 3.64PCLDD515 pKa = 3.5NPAYY519 pKa = 9.14PVPQSTGVTHH529 pKa = 7.11PLFAGAGGITAQVGPSGLTARR550 pKa = 11.84TIRR553 pKa = 11.84ALCASPPKK561 pKa = 10.62AQTSQCAFLGGDD573 pKa = 3.03RR574 pKa = 11.84SEE576 pKa = 4.09EE577 pKa = 4.14EE578 pKa = 3.98KK579 pKa = 10.83RR580 pKa = 11.84KK581 pKa = 8.29EE582 pKa = 4.12TQEE585 pKa = 4.05TSSEE589 pKa = 4.11SSITSAEE596 pKa = 4.07SSTEE600 pKa = 3.94GDD602 pKa = 3.09GSTDD606 pKa = 3.12EE607 pKa = 4.12EE608 pKa = 4.11EE609 pKa = 3.9RR610 pKa = 11.84RR611 pKa = 11.84EE612 pKa = 3.76RR613 pKa = 11.84RR614 pKa = 11.84RR615 pKa = 11.84RR616 pKa = 11.84RR617 pKa = 11.84RR618 pKa = 11.84KK619 pKa = 8.84RR620 pKa = 11.84QLKK623 pKa = 10.15LFYY626 pKa = 10.37QRR628 pKa = 11.84LTDD631 pKa = 4.24RR632 pKa = 11.84YY633 pKa = 9.55LEE635 pKa = 4.11HH636 pKa = 6.71KK637 pKa = 10.16RR638 pKa = 11.84RR639 pKa = 11.84RR640 pKa = 11.84LSEE643 pKa = 3.83
MM1 pKa = 8.06RR2 pKa = 11.84FRR4 pKa = 11.84PIRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84YY14 pKa = 9.34RR15 pKa = 11.84KK16 pKa = 9.55RR17 pKa = 11.84RR18 pKa = 11.84WGWRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 9.16YY26 pKa = 8.7RR27 pKa = 11.84HH28 pKa = 6.54HH29 pKa = 5.95YY30 pKa = 9.16RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84PWRR36 pKa = 11.84RR37 pKa = 11.84WRR39 pKa = 11.84VRR41 pKa = 11.84RR42 pKa = 11.84WRR44 pKa = 11.84RR45 pKa = 11.84SVFRR49 pKa = 11.84RR50 pKa = 11.84GGRR53 pKa = 11.84RR54 pKa = 11.84ARR56 pKa = 11.84PYY58 pKa = 10.18RR59 pKa = 11.84ISAFNPRR66 pKa = 11.84VMRR69 pKa = 11.84RR70 pKa = 11.84IVIQGWWPLIQCLPGQEE87 pKa = 4.09HH88 pKa = 6.75LRR90 pKa = 11.84YY91 pKa = 9.65RR92 pKa = 11.84PLNWDD97 pKa = 3.3IEE99 pKa = 4.48KK100 pKa = 10.44EE101 pKa = 4.22WVVEE105 pKa = 3.97KK106 pKa = 10.95DD107 pKa = 3.44KK108 pKa = 11.52EE109 pKa = 4.14PDD111 pKa = 2.97KK112 pKa = 10.85MGYY115 pKa = 9.11LVQYY119 pKa = 10.1GGGWGSGHH127 pKa = 6.31ITLEE131 pKa = 4.07DD132 pKa = 4.17LFNEE136 pKa = 4.14HH137 pKa = 5.92QKK139 pKa = 8.42WRR141 pKa = 11.84NMWSKK146 pKa = 11.53SNDD149 pKa = 2.85GMDD152 pKa = 3.68LVRR155 pKa = 11.84YY156 pKa = 7.33FGCKK160 pKa = 8.81IYY162 pKa = 10.43LYY164 pKa = 9.82PLEE167 pKa = 5.55DD168 pKa = 2.88VDD170 pKa = 4.93YY171 pKa = 9.82WFWWDD176 pKa = 3.06TDD178 pKa = 3.77FKK180 pKa = 11.64NNYY183 pKa = 6.99EE184 pKa = 4.07DD185 pKa = 5.33RR186 pKa = 11.84IKK188 pKa = 10.44EE189 pKa = 3.98YY190 pKa = 10.45CQPSVMLMAKK200 pKa = 8.32RR201 pKa = 11.84TKK203 pKa = 10.03IVVARR208 pKa = 11.84KK209 pKa = 8.72RR210 pKa = 11.84AQHH213 pKa = 5.56RR214 pKa = 11.84RR215 pKa = 11.84RR216 pKa = 11.84VKK218 pKa = 10.46KK219 pKa = 10.42IFIPPPSRR227 pKa = 11.84DD228 pKa = 3.04TTQWKK233 pKa = 9.0FQTEE237 pKa = 4.5FCKK240 pKa = 10.7QPLFTWAAGLIEE252 pKa = 4.38LQRR255 pKa = 11.84PFDD258 pKa = 4.05FNGGFRR264 pKa = 11.84NAWWLEE270 pKa = 3.49PRR272 pKa = 11.84NEE274 pKa = 4.27AGEE277 pKa = 4.1MDD279 pKa = 5.56YY280 pKa = 9.86ITLWGRR286 pKa = 11.84VPPQGDD292 pKa = 3.19TDD294 pKa = 3.81MPDD297 pKa = 3.28PNTFKK302 pKa = 8.72TTTQNKK308 pKa = 9.47NYY310 pKa = 9.93SIQPGKK316 pKa = 10.05EE317 pKa = 3.79NNVYY321 pKa = 10.44PIMAYY326 pKa = 9.16IRR328 pKa = 11.84QEE330 pKa = 3.79NQQPVVNWCVCYY342 pKa = 10.89NKK344 pKa = 10.42TIDD347 pKa = 3.15RR348 pKa = 11.84WRR350 pKa = 11.84KK351 pKa = 6.27TQAATLKK358 pKa = 10.18IGNLRR363 pKa = 11.84GLCVRR368 pKa = 11.84EE369 pKa = 4.55LSRR372 pKa = 11.84QEE374 pKa = 3.59MTYY377 pKa = 10.05KK378 pKa = 10.02WKK380 pKa = 10.57QGEE383 pKa = 4.03FSKK386 pKa = 10.55AYY388 pKa = 9.41HH389 pKa = 6.63DD390 pKa = 3.18RR391 pKa = 11.84WGYY394 pKa = 7.7NTYY397 pKa = 10.33FVTIDD402 pKa = 3.37GRR404 pKa = 11.84KK405 pKa = 7.74GTEE408 pKa = 3.89NPTVQIWNFNDD419 pKa = 3.9KK420 pKa = 9.38ITVQSGHH427 pKa = 6.48ILNFAFNIQKK437 pKa = 10.88SNDD440 pKa = 3.41NLKK443 pKa = 11.03DD444 pKa = 3.35SDD446 pKa = 3.89FGKK449 pKa = 10.42IGLPKK454 pKa = 10.24SPHH457 pKa = 6.57DD458 pKa = 4.62LDD460 pKa = 5.33FGHH463 pKa = 7.24KK464 pKa = 9.92NRR466 pKa = 11.84FGPFVVKK473 pKa = 10.77NEE475 pKa = 3.89PVEE478 pKa = 4.23FQLNPPRR485 pKa = 11.84AINVWVRR492 pKa = 11.84YY493 pKa = 10.0KK494 pKa = 10.85FFFQFGGEE502 pKa = 4.11YY503 pKa = 10.14QPPTGIRR510 pKa = 11.84DD511 pKa = 3.64PCLDD515 pKa = 3.5NPAYY519 pKa = 9.14PVPQSTGVTHH529 pKa = 7.11PLFAGAGGITAQVGPSGLTARR550 pKa = 11.84TIRR553 pKa = 11.84ALCASPPKK561 pKa = 10.62AQTSQCAFLGGDD573 pKa = 3.03RR574 pKa = 11.84SEE576 pKa = 4.09EE577 pKa = 4.14EE578 pKa = 3.98KK579 pKa = 10.83RR580 pKa = 11.84KK581 pKa = 8.29EE582 pKa = 4.12TQEE585 pKa = 4.05TSSEE589 pKa = 4.11SSITSAEE596 pKa = 4.07SSTEE600 pKa = 3.94GDD602 pKa = 3.09GSTDD606 pKa = 3.12EE607 pKa = 4.12EE608 pKa = 4.11EE609 pKa = 3.9RR610 pKa = 11.84RR611 pKa = 11.84EE612 pKa = 3.76RR613 pKa = 11.84RR614 pKa = 11.84RR615 pKa = 11.84RR616 pKa = 11.84RR617 pKa = 11.84RR618 pKa = 11.84KK619 pKa = 8.84RR620 pKa = 11.84QLKK623 pKa = 10.15LFYY626 pKa = 10.37QRR628 pKa = 11.84LTDD631 pKa = 4.24RR632 pKa = 11.84YY633 pKa = 9.55LEE635 pKa = 4.11HH636 pKa = 6.71KK637 pKa = 10.16RR638 pKa = 11.84RR639 pKa = 11.84RR640 pKa = 11.84LSEE643 pKa = 3.83
Molecular weight: 77.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
715 |
72 |
643 |
357.5 |
42.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.035 ± 4.19 | 1.818 ± 0.961 |
5.874 ± 3.279 | 6.014 ± 0.381 |
4.615 ± 1.32 | 7.273 ± 1.57 |
1.958 ± 0.335 | 4.615 ± 0.184 |
5.734 ± 2.346 | 5.874 ± 1.574 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.678 ± 0.118 | 4.755 ± 0.809 |
5.734 ± 1.778 | 4.895 ± 0.866 |
11.748 ± 3.102 | 3.916 ± 1.602 |
5.594 ± 0.552 | 4.476 ± 0.695 |
4.056 ± 0.045 | 4.336 ± 0.069 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
