
Arthrobacter phage Mendel
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; Anjalivirus; Arthobacter virus Mendel
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
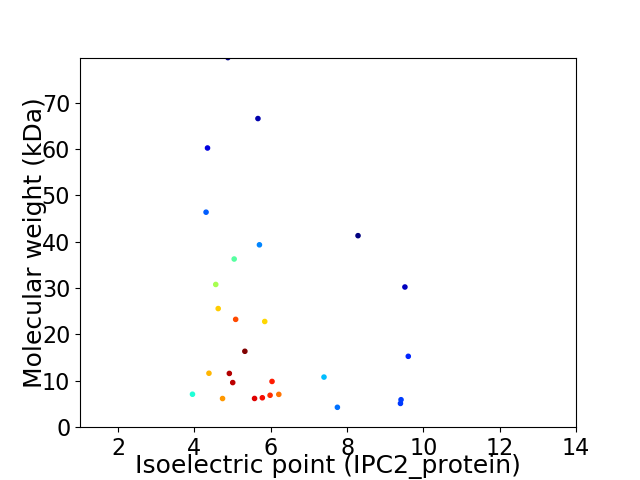
Virtual 2D-PAGE plot for 28 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G3M0K4|A0A3G3M0K4_9CAUD Uncharacterized protein OS=Arthrobacter phage Mendel OX=2484218 GN=1 PE=4 SV=1
MM1 pKa = 6.74VTTATGVFPYY11 pKa = 10.47RR12 pKa = 11.84LQPLNNISAFTYY24 pKa = 8.76RR25 pKa = 11.84TGVTYY30 pKa = 11.08AEE32 pKa = 4.22MLEE35 pKa = 4.03EE36 pKa = 3.91LRR38 pKa = 11.84VYY40 pKa = 10.71INDD43 pKa = 3.43TMRR46 pKa = 11.84PEE48 pKa = 4.2FNSVILSVLNDD59 pKa = 3.74FQAGIEE65 pKa = 4.1NAEE68 pKa = 4.08TRR70 pKa = 11.84VVQSEE75 pKa = 4.84TEE77 pKa = 4.23FTALMDD83 pKa = 4.2DD84 pKa = 3.0ARR86 pKa = 11.84AYY88 pKa = 11.03VDD90 pKa = 4.69ASVQFINNKK99 pKa = 8.31TGQAQIQRR107 pKa = 11.84VTLTAPYY114 pKa = 9.51TVTIDD119 pKa = 3.7PLWPNNHH126 pKa = 7.57PINIVLTQDD135 pKa = 3.04AAGGHH140 pKa = 5.81AVTWDD145 pKa = 3.55SEE147 pKa = 4.58DD148 pKa = 3.38INGSVSVSTAPNAEE162 pKa = 3.76TDD164 pKa = 3.64FWLVPEE170 pKa = 5.39GDD172 pKa = 4.18GTWTAVQYY180 pKa = 8.48VTAVQLAGEE189 pKa = 4.33VATINTALGTKK200 pKa = 10.05ANSSSLATKK209 pKa = 10.12ADD211 pKa = 3.31KK212 pKa = 10.23ATTITTSKK220 pKa = 9.66PLYY223 pKa = 10.09GIAGVIRR230 pKa = 11.84NTGAGWDD237 pKa = 3.9VINDD241 pKa = 3.87AGHH244 pKa = 6.0EE245 pKa = 4.32PINIDD250 pKa = 3.46NVVSDD255 pKa = 4.15GDD257 pKa = 4.42KK258 pKa = 9.85ITINYY263 pKa = 6.79GTLGASKK270 pKa = 10.47VGTFIVTPDD279 pKa = 3.46EE280 pKa = 4.32KK281 pKa = 11.22LNEE284 pKa = 3.91QGFQVGASVGPTSTSIYY301 pKa = 9.52LSRR304 pKa = 11.84SIPSYY309 pKa = 10.62SDD311 pKa = 3.2YY312 pKa = 11.4VAYY315 pKa = 9.69TSGAFVSANGVFTCTWNATDD335 pKa = 5.22GYY337 pKa = 10.64LQVTHH342 pKa = 6.02PTIIGSALNVSLAGRR357 pKa = 11.84PGTALNYY364 pKa = 8.54TPVVAGDD371 pKa = 3.86LTTGSTFIAIEE382 pKa = 3.97FRR384 pKa = 11.84DD385 pKa = 3.56AAGNQVMTPDD395 pKa = 2.86TNMRR399 pKa = 11.84VFVTHH404 pKa = 6.71GGGFVPSVDD413 pKa = 3.29PSTVTTATYY422 pKa = 10.09PNSNLWIYY430 pKa = 11.44AVMQDD435 pKa = 3.2
MM1 pKa = 6.74VTTATGVFPYY11 pKa = 10.47RR12 pKa = 11.84LQPLNNISAFTYY24 pKa = 8.76RR25 pKa = 11.84TGVTYY30 pKa = 11.08AEE32 pKa = 4.22MLEE35 pKa = 4.03EE36 pKa = 3.91LRR38 pKa = 11.84VYY40 pKa = 10.71INDD43 pKa = 3.43TMRR46 pKa = 11.84PEE48 pKa = 4.2FNSVILSVLNDD59 pKa = 3.74FQAGIEE65 pKa = 4.1NAEE68 pKa = 4.08TRR70 pKa = 11.84VVQSEE75 pKa = 4.84TEE77 pKa = 4.23FTALMDD83 pKa = 4.2DD84 pKa = 3.0ARR86 pKa = 11.84AYY88 pKa = 11.03VDD90 pKa = 4.69ASVQFINNKK99 pKa = 8.31TGQAQIQRR107 pKa = 11.84VTLTAPYY114 pKa = 9.51TVTIDD119 pKa = 3.7PLWPNNHH126 pKa = 7.57PINIVLTQDD135 pKa = 3.04AAGGHH140 pKa = 5.81AVTWDD145 pKa = 3.55SEE147 pKa = 4.58DD148 pKa = 3.38INGSVSVSTAPNAEE162 pKa = 3.76TDD164 pKa = 3.64FWLVPEE170 pKa = 5.39GDD172 pKa = 4.18GTWTAVQYY180 pKa = 8.48VTAVQLAGEE189 pKa = 4.33VATINTALGTKK200 pKa = 10.05ANSSSLATKK209 pKa = 10.12ADD211 pKa = 3.31KK212 pKa = 10.23ATTITTSKK220 pKa = 9.66PLYY223 pKa = 10.09GIAGVIRR230 pKa = 11.84NTGAGWDD237 pKa = 3.9VINDD241 pKa = 3.87AGHH244 pKa = 6.0EE245 pKa = 4.32PINIDD250 pKa = 3.46NVVSDD255 pKa = 4.15GDD257 pKa = 4.42KK258 pKa = 9.85ITINYY263 pKa = 6.79GTLGASKK270 pKa = 10.47VGTFIVTPDD279 pKa = 3.46EE280 pKa = 4.32KK281 pKa = 11.22LNEE284 pKa = 3.91QGFQVGASVGPTSTSIYY301 pKa = 9.52LSRR304 pKa = 11.84SIPSYY309 pKa = 10.62SDD311 pKa = 3.2YY312 pKa = 11.4VAYY315 pKa = 9.69TSGAFVSANGVFTCTWNATDD335 pKa = 5.22GYY337 pKa = 10.64LQVTHH342 pKa = 6.02PTIIGSALNVSLAGRR357 pKa = 11.84PGTALNYY364 pKa = 8.54TPVVAGDD371 pKa = 3.86LTTGSTFIAIEE382 pKa = 3.97FRR384 pKa = 11.84DD385 pKa = 3.56AAGNQVMTPDD395 pKa = 2.86TNMRR399 pKa = 11.84VFVTHH404 pKa = 6.71GGGFVPSVDD413 pKa = 3.29PSTVTTATYY422 pKa = 10.09PNSNLWIYY430 pKa = 11.44AVMQDD435 pKa = 3.2
Molecular weight: 46.38 kDa
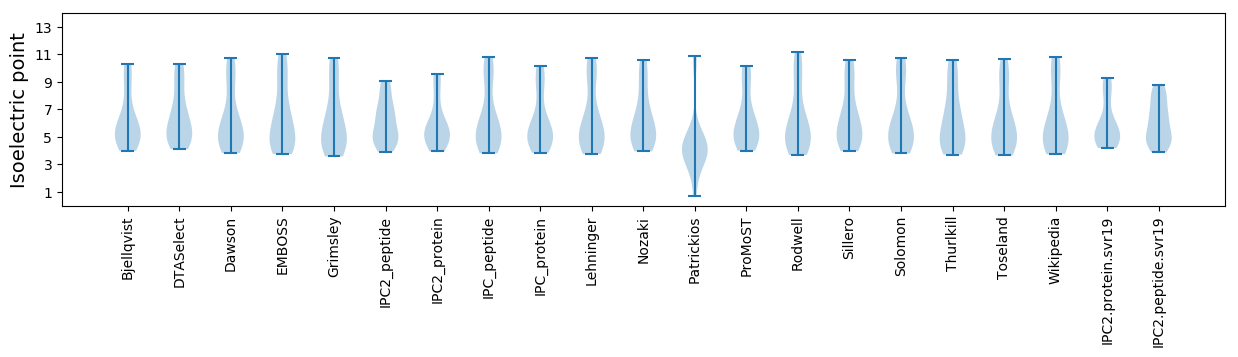
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G3M0F4|A0A3G3M0F4_9CAUD Terminase OS=Arthrobacter phage Mendel OX=2484218 GN=18 PE=4 SV=1
MM1 pKa = 7.92GDD3 pKa = 3.42TMHH6 pKa = 7.58KK7 pKa = 9.8ATATPLTQLKK17 pKa = 9.81EE18 pKa = 3.77RR19 pKa = 11.84LARR22 pKa = 11.84LGRR25 pKa = 11.84NNLHH29 pKa = 5.97RR30 pKa = 11.84CYY32 pKa = 10.8GKK34 pKa = 10.26RR35 pKa = 11.84CADD38 pKa = 3.68HH39 pKa = 6.36TNNWYY44 pKa = 10.18AWATGTAKK52 pKa = 10.66
MM1 pKa = 7.92GDD3 pKa = 3.42TMHH6 pKa = 7.58KK7 pKa = 9.8ATATPLTQLKK17 pKa = 9.81EE18 pKa = 3.77RR19 pKa = 11.84LARR22 pKa = 11.84LGRR25 pKa = 11.84NNLHH29 pKa = 5.97RR30 pKa = 11.84CYY32 pKa = 10.8GKK34 pKa = 10.26RR35 pKa = 11.84CADD38 pKa = 3.68HH39 pKa = 6.36TNNWYY44 pKa = 10.18AWATGTAKK52 pKa = 10.66
Molecular weight: 5.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5872 |
40 |
733 |
209.7 |
22.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.746 ± 0.465 | 0.613 ± 0.125 |
6.233 ± 0.331 | 5.109 ± 0.5 |
3.883 ± 0.411 | 8.345 ± 0.561 |
1.839 ± 0.253 | 5.381 ± 0.311 |
4.155 ± 0.478 | 7.084 ± 0.414 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.265 ± 0.191 | 5.211 ± 0.471 |
5.126 ± 0.349 | 3.815 ± 0.324 |
4.905 ± 0.478 | 4.973 ± 0.256 |
8.328 ± 0.651 | 6.574 ± 0.506 |
1.686 ± 0.113 | 3.73 ± 0.361 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
