
Cynomolgus macaque cytomegalovirus strain Mauritius
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Betaherpesvirinae; Cytomegalovirus; Macacine betaherpesvirus 3
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins
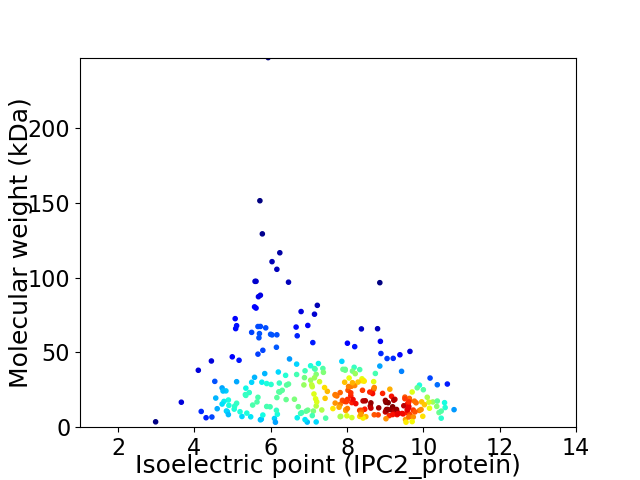
Virtual 2D-PAGE plot for 290 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1H0C2|A0A0K1H0C2_9BETA Uncharacterized protein OS=Cynomolgus macaque cytomegalovirus strain Mauritius OX=1690255 GN=Cy20 PE=4 SV=1
MM1 pKa = 7.5RR2 pKa = 11.84WVAMRR7 pKa = 11.84WLLLIVATSAKK18 pKa = 10.29ASSTTAGNTSTPTASTITSTVTTASTTNMSTPSPASSPATTNTTTSSPSTKK69 pKa = 10.33SSTSSTNTTITTTGTASATTPKK91 pKa = 10.4SSTSVTSSPANSTSTSTKK109 pKa = 9.78PSTTNTSSPITTSSPASNTSTTTSSTSTVNGTNSTTEE146 pKa = 4.13TNTTSLPDD154 pKa = 3.27TTADD158 pKa = 3.45MNVTTTEE165 pKa = 4.5PYY167 pKa = 10.96NSTGYY172 pKa = 11.03EE173 pKa = 3.7NVTINITTPSPYY185 pKa = 10.0EE186 pKa = 3.61ALQIVDD192 pKa = 4.79LCNEE196 pKa = 4.43TISIVFKK203 pKa = 11.14DD204 pKa = 3.61PGEE207 pKa = 4.06EE208 pKa = 4.2DD209 pKa = 3.46EE210 pKa = 5.09SSEE213 pKa = 3.95TTEE216 pKa = 4.2YY217 pKa = 11.17SDD219 pKa = 4.21EE220 pKa = 4.3EE221 pKa = 4.3PVASSNEE228 pKa = 3.83DD229 pKa = 3.2DD230 pKa = 3.48STYY233 pKa = 10.3FPQSPGYY240 pKa = 7.88TLTYY244 pKa = 9.43DD245 pKa = 3.78TEE247 pKa = 4.17DD248 pKa = 3.27TIYY251 pKa = 10.65FQATCDD257 pKa = 4.08RR258 pKa = 11.84NDD260 pKa = 3.33TYY262 pKa = 11.56NITSCDD268 pKa = 3.68YY269 pKa = 10.21TNKK272 pKa = 10.2SVNSWSTVTSVSFFPPTLTPCHH294 pKa = 6.56KK295 pKa = 9.56PVAIIKK301 pKa = 9.88IGNDD305 pKa = 3.25SLVVSASATSNLVDD319 pKa = 5.76AIYY322 pKa = 10.99KK323 pKa = 10.49LLGLPDD329 pKa = 3.78VNSDD333 pKa = 4.72FINQLGRR340 pKa = 11.84YY341 pKa = 8.7HH342 pKa = 7.95PITLQGQIEE351 pKa = 4.19YY352 pKa = 9.97RR353 pKa = 11.84DD354 pKa = 3.61WYY356 pKa = 7.62TTEE359 pKa = 3.67
MM1 pKa = 7.5RR2 pKa = 11.84WVAMRR7 pKa = 11.84WLLLIVATSAKK18 pKa = 10.29ASSTTAGNTSTPTASTITSTVTTASTTNMSTPSPASSPATTNTTTSSPSTKK69 pKa = 10.33SSTSSTNTTITTTGTASATTPKK91 pKa = 10.4SSTSVTSSPANSTSTSTKK109 pKa = 9.78PSTTNTSSPITTSSPASNTSTTTSSTSTVNGTNSTTEE146 pKa = 4.13TNTTSLPDD154 pKa = 3.27TTADD158 pKa = 3.45MNVTTTEE165 pKa = 4.5PYY167 pKa = 10.96NSTGYY172 pKa = 11.03EE173 pKa = 3.7NVTINITTPSPYY185 pKa = 10.0EE186 pKa = 3.61ALQIVDD192 pKa = 4.79LCNEE196 pKa = 4.43TISIVFKK203 pKa = 11.14DD204 pKa = 3.61PGEE207 pKa = 4.06EE208 pKa = 4.2DD209 pKa = 3.46EE210 pKa = 5.09SSEE213 pKa = 3.95TTEE216 pKa = 4.2YY217 pKa = 11.17SDD219 pKa = 4.21EE220 pKa = 4.3EE221 pKa = 4.3PVASSNEE228 pKa = 3.83DD229 pKa = 3.2DD230 pKa = 3.48STYY233 pKa = 10.3FPQSPGYY240 pKa = 7.88TLTYY244 pKa = 9.43DD245 pKa = 3.78TEE247 pKa = 4.17DD248 pKa = 3.27TIYY251 pKa = 10.65FQATCDD257 pKa = 4.08RR258 pKa = 11.84NDD260 pKa = 3.33TYY262 pKa = 11.56NITSCDD268 pKa = 3.68YY269 pKa = 10.21TNKK272 pKa = 10.2SVNSWSTVTSVSFFPPTLTPCHH294 pKa = 6.56KK295 pKa = 9.56PVAIIKK301 pKa = 9.88IGNDD305 pKa = 3.25SLVVSASATSNLVDD319 pKa = 5.76AIYY322 pKa = 10.99KK323 pKa = 10.49LLGLPDD329 pKa = 3.78VNSDD333 pKa = 4.72FINQLGRR340 pKa = 11.84YY341 pKa = 8.7HH342 pKa = 7.95PITLQGQIEE351 pKa = 4.19YY352 pKa = 9.97RR353 pKa = 11.84DD354 pKa = 3.61WYY356 pKa = 7.62TTEE359 pKa = 3.67
Molecular weight: 38.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1H019|A0A0K1H019_9BETA Uncharacterized protein OS=Cynomolgus macaque cytomegalovirus strain Mauritius OX=1690255 GN=Cy83 PE=4 SV=1
MM1 pKa = 6.66VQIHH5 pKa = 6.22EE6 pKa = 4.41SCGTVRR12 pKa = 11.84GPGRR16 pKa = 11.84SRR18 pKa = 11.84ALHH21 pKa = 6.12RR22 pKa = 11.84SVALLPLSDD31 pKa = 4.37ASGDD35 pKa = 3.15RR36 pKa = 11.84HH37 pKa = 5.49GRR39 pKa = 11.84CRR41 pKa = 11.84QDD43 pKa = 2.96VQYY46 pKa = 11.06SGAGCEE52 pKa = 4.09PKK54 pKa = 10.18LRR56 pKa = 11.84HH57 pKa = 5.84YY58 pKa = 10.43RR59 pKa = 11.84DD60 pKa = 3.09HH61 pKa = 7.32RR62 pKa = 11.84NRR64 pKa = 11.84RR65 pKa = 11.84PEE67 pKa = 3.99PQRR70 pKa = 11.84DD71 pKa = 4.05SQPHH75 pKa = 5.74ALGAGEE81 pKa = 5.38DD82 pKa = 3.84DD83 pKa = 5.75LSRR86 pKa = 11.84LWFQQQTCASGRR98 pKa = 11.84GRR100 pKa = 11.84QRR102 pKa = 11.84YY103 pKa = 9.11SEE105 pKa = 4.53AVPDD109 pKa = 3.59LRR111 pKa = 11.84VARR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 9.4HH117 pKa = 5.72HH118 pKa = 5.79STSASEE124 pKa = 4.1RR125 pKa = 11.84SLDD128 pKa = 3.5LLAGHH133 pKa = 6.74RR134 pKa = 11.84RR135 pKa = 11.84HH136 pKa = 6.0YY137 pKa = 11.1GKK139 pKa = 9.87ILKK142 pKa = 10.05YY143 pKa = 10.43VGTKK147 pKa = 10.17GVSS150 pKa = 2.92
MM1 pKa = 6.66VQIHH5 pKa = 6.22EE6 pKa = 4.41SCGTVRR12 pKa = 11.84GPGRR16 pKa = 11.84SRR18 pKa = 11.84ALHH21 pKa = 6.12RR22 pKa = 11.84SVALLPLSDD31 pKa = 4.37ASGDD35 pKa = 3.15RR36 pKa = 11.84HH37 pKa = 5.49GRR39 pKa = 11.84CRR41 pKa = 11.84QDD43 pKa = 2.96VQYY46 pKa = 11.06SGAGCEE52 pKa = 4.09PKK54 pKa = 10.18LRR56 pKa = 11.84HH57 pKa = 5.84YY58 pKa = 10.43RR59 pKa = 11.84DD60 pKa = 3.09HH61 pKa = 7.32RR62 pKa = 11.84NRR64 pKa = 11.84RR65 pKa = 11.84PEE67 pKa = 3.99PQRR70 pKa = 11.84DD71 pKa = 4.05SQPHH75 pKa = 5.74ALGAGEE81 pKa = 5.38DD82 pKa = 3.84DD83 pKa = 5.75LSRR86 pKa = 11.84LWFQQQTCASGRR98 pKa = 11.84GRR100 pKa = 11.84QRR102 pKa = 11.84YY103 pKa = 9.11SEE105 pKa = 4.53AVPDD109 pKa = 3.59LRR111 pKa = 11.84VARR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 9.4HH117 pKa = 5.72HH118 pKa = 5.79STSASEE124 pKa = 4.1RR125 pKa = 11.84SLDD128 pKa = 3.5LLAGHH133 pKa = 6.74RR134 pKa = 11.84RR135 pKa = 11.84HH136 pKa = 6.0YY137 pKa = 11.1GKK139 pKa = 9.87ILKK142 pKa = 10.05YY143 pKa = 10.43VGTKK147 pKa = 10.17GVSS150 pKa = 2.92
Molecular weight: 16.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
72865 |
29 |
2179 |
251.3 |
28.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.763 ± 0.146 | 2.743 ± 0.104 |
4.451 ± 0.123 | 4.857 ± 0.135 |
4.116 ± 0.101 | 4.916 ± 0.158 |
3.318 ± 0.114 | 5.096 ± 0.139 |
3.663 ± 0.131 | 9.901 ± 0.171 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.665 ± 0.078 | 3.996 ± 0.113 |
5.754 ± 0.169 | 3.94 ± 0.134 |
6.903 ± 0.198 | 7.954 ± 0.154 |
6.986 ± 0.236 | 7.08 ± 0.148 |
1.433 ± 0.082 | 3.464 ± 0.098 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
