
Jatropha mosaic India virus-[Lucknow]
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus; Jatropha mosaic India virus
Average proteome isoelectric point is 8.0
Get precalculated fractions of proteins
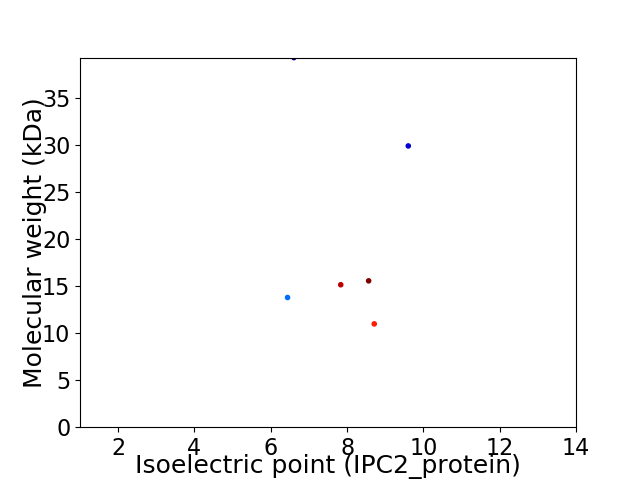
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2FZU7|E2FZU7_9GEMI Capsid protein OS=Jatropha mosaic India virus-[Lucknow] OX=569949 GN=AV1 PE=3 SV=1
MM1 pKa = 8.06WDD3 pKa = 3.35PLLNEE8 pKa = 4.99FPDD11 pKa = 3.86SVHH14 pKa = 6.81GFRR17 pKa = 11.84CMLALKK23 pKa = 9.76YY24 pKa = 10.16LQLVEE29 pKa = 4.41STYY32 pKa = 11.41SPDD35 pKa = 3.14TLGYY39 pKa = 10.84DD40 pKa = 4.83LIRR43 pKa = 11.84DD44 pKa = 4.71LISVIRR50 pKa = 11.84ARR52 pKa = 11.84SYY54 pKa = 10.28VEE56 pKa = 3.27ATRR59 pKa = 11.84RR60 pKa = 11.84YY61 pKa = 8.39HH62 pKa = 6.31HH63 pKa = 6.62FNSRR67 pKa = 11.84LEE69 pKa = 4.21GTSPSDD75 pKa = 3.47LRR77 pKa = 11.84QLIQQPCFCLHH88 pKa = 6.9CPRR91 pKa = 11.84HH92 pKa = 5.61QKK94 pKa = 9.39TSLDD98 pKa = 3.79KK99 pKa = 10.61QAHH102 pKa = 5.06EE103 pKa = 5.14SEE105 pKa = 4.26AQMVQDD111 pKa = 3.71VQKK114 pKa = 10.17PRR116 pKa = 11.84CSS118 pKa = 3.2
MM1 pKa = 8.06WDD3 pKa = 3.35PLLNEE8 pKa = 4.99FPDD11 pKa = 3.86SVHH14 pKa = 6.81GFRR17 pKa = 11.84CMLALKK23 pKa = 9.76YY24 pKa = 10.16LQLVEE29 pKa = 4.41STYY32 pKa = 11.41SPDD35 pKa = 3.14TLGYY39 pKa = 10.84DD40 pKa = 4.83LIRR43 pKa = 11.84DD44 pKa = 4.71LISVIRR50 pKa = 11.84ARR52 pKa = 11.84SYY54 pKa = 10.28VEE56 pKa = 3.27ATRR59 pKa = 11.84RR60 pKa = 11.84YY61 pKa = 8.39HH62 pKa = 6.31HH63 pKa = 6.62FNSRR67 pKa = 11.84LEE69 pKa = 4.21GTSPSDD75 pKa = 3.47LRR77 pKa = 11.84QLIQQPCFCLHH88 pKa = 6.9CPRR91 pKa = 11.84HH92 pKa = 5.61QKK94 pKa = 9.39TSLDD98 pKa = 3.79KK99 pKa = 10.61QAHH102 pKa = 5.06EE103 pKa = 5.14SEE105 pKa = 4.26AQMVQDD111 pKa = 3.71VQKK114 pKa = 10.17PRR116 pKa = 11.84CSS118 pKa = 3.2
Molecular weight: 13.79 kDa
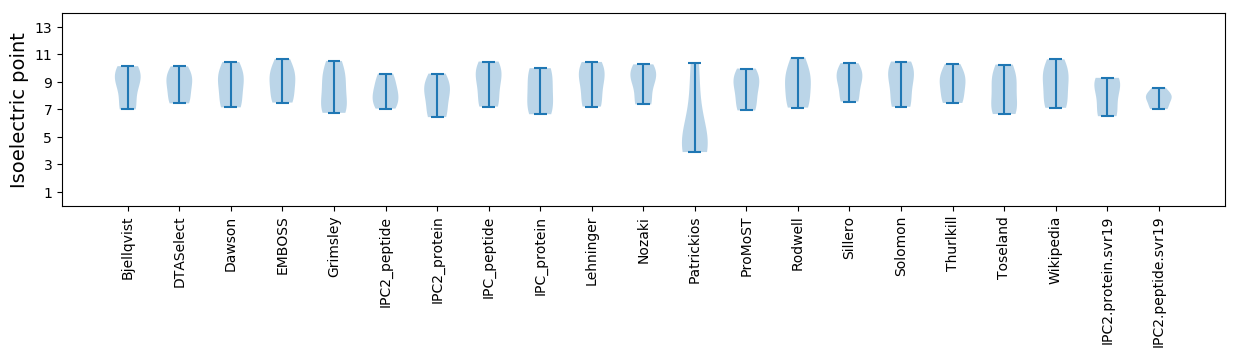
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2FZU8|E2FZU8_9GEMI Replication enhancer OS=Jatropha mosaic India virus-[Lucknow] OX=569949 GN=AC3 PE=3 SV=1
MM1 pKa = 7.7SKK3 pKa = 10.44RR4 pKa = 11.84PGDD7 pKa = 3.89IIISTPASKK16 pKa = 10.31VRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LIFDD24 pKa = 3.31SLYY27 pKa = 10.61SSRR30 pKa = 11.84ASVFTVRR37 pKa = 11.84VTKK40 pKa = 10.46RR41 pKa = 11.84QAWTNRR47 pKa = 11.84PMNRR51 pKa = 11.84KK52 pKa = 7.93PRR54 pKa = 11.84WYY56 pKa = 10.73RR57 pKa = 11.84MFRR60 pKa = 11.84SPDD63 pKa = 3.23VPRR66 pKa = 11.84GCEE69 pKa = 4.34GPCKK73 pKa = 10.01VQSFEE78 pKa = 4.02SRR80 pKa = 11.84HH81 pKa = 5.54DD82 pKa = 3.51VVHH85 pKa = 6.75IGKK88 pKa = 9.41VMCISDD94 pKa = 3.53VTRR97 pKa = 11.84GVGLTHH103 pKa = 7.14RR104 pKa = 11.84VGKK107 pKa = 9.66RR108 pKa = 11.84FCVKK112 pKa = 9.92SVYY115 pKa = 10.24ILGKK119 pKa = 9.73VWMDD123 pKa = 3.27EE124 pKa = 4.06NIKK127 pKa = 9.31TKK129 pKa = 10.6NHH131 pKa = 5.78TNSVMFFLVRR141 pKa = 11.84DD142 pKa = 3.84RR143 pKa = 11.84RR144 pKa = 11.84PVDD147 pKa = 3.13KK148 pKa = 10.28PQDD151 pKa = 3.56FGEE154 pKa = 4.33VFNMFDD160 pKa = 4.29NEE162 pKa = 4.11PSTATVKK169 pKa = 10.61NMHH172 pKa = 7.0RR173 pKa = 11.84DD174 pKa = 3.31RR175 pKa = 11.84YY176 pKa = 9.22QVLRR180 pKa = 11.84KK181 pKa = 9.45WSATVTGGQYY191 pKa = 10.85ASKK194 pKa = 9.9EE195 pKa = 3.88QALVRR200 pKa = 11.84RR201 pKa = 11.84FFRR204 pKa = 11.84VNNYY208 pKa = 7.82VVYY211 pKa = 9.96NQQEE215 pKa = 3.8AGKK218 pKa = 10.1YY219 pKa = 8.23EE220 pKa = 4.02NHH222 pKa = 6.47TEE224 pKa = 3.99NALMLYY230 pKa = 7.52MACTHH235 pKa = 7.06ASNPVYY241 pKa = 9.86ATLKK245 pKa = 9.47IRR247 pKa = 11.84IYY249 pKa = 10.67FYY251 pKa = 11.26DD252 pKa = 3.51SVSNN256 pKa = 3.9
MM1 pKa = 7.7SKK3 pKa = 10.44RR4 pKa = 11.84PGDD7 pKa = 3.89IIISTPASKK16 pKa = 10.31VRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LIFDD24 pKa = 3.31SLYY27 pKa = 10.61SSRR30 pKa = 11.84ASVFTVRR37 pKa = 11.84VTKK40 pKa = 10.46RR41 pKa = 11.84QAWTNRR47 pKa = 11.84PMNRR51 pKa = 11.84KK52 pKa = 7.93PRR54 pKa = 11.84WYY56 pKa = 10.73RR57 pKa = 11.84MFRR60 pKa = 11.84SPDD63 pKa = 3.23VPRR66 pKa = 11.84GCEE69 pKa = 4.34GPCKK73 pKa = 10.01VQSFEE78 pKa = 4.02SRR80 pKa = 11.84HH81 pKa = 5.54DD82 pKa = 3.51VVHH85 pKa = 6.75IGKK88 pKa = 9.41VMCISDD94 pKa = 3.53VTRR97 pKa = 11.84GVGLTHH103 pKa = 7.14RR104 pKa = 11.84VGKK107 pKa = 9.66RR108 pKa = 11.84FCVKK112 pKa = 9.92SVYY115 pKa = 10.24ILGKK119 pKa = 9.73VWMDD123 pKa = 3.27EE124 pKa = 4.06NIKK127 pKa = 9.31TKK129 pKa = 10.6NHH131 pKa = 5.78TNSVMFFLVRR141 pKa = 11.84DD142 pKa = 3.84RR143 pKa = 11.84RR144 pKa = 11.84PVDD147 pKa = 3.13KK148 pKa = 10.28PQDD151 pKa = 3.56FGEE154 pKa = 4.33VFNMFDD160 pKa = 4.29NEE162 pKa = 4.11PSTATVKK169 pKa = 10.61NMHH172 pKa = 7.0RR173 pKa = 11.84DD174 pKa = 3.31RR175 pKa = 11.84YY176 pKa = 9.22QVLRR180 pKa = 11.84KK181 pKa = 9.45WSATVTGGQYY191 pKa = 10.85ASKK194 pKa = 9.9EE195 pKa = 3.88QALVRR200 pKa = 11.84RR201 pKa = 11.84FFRR204 pKa = 11.84VNNYY208 pKa = 7.82VVYY211 pKa = 9.96NQQEE215 pKa = 3.8AGKK218 pKa = 10.1YY219 pKa = 8.23EE220 pKa = 4.02NHH222 pKa = 6.47TEE224 pKa = 3.99NALMLYY230 pKa = 7.52MACTHH235 pKa = 7.06ASNPVYY241 pKa = 9.86ATLKK245 pKa = 9.47IRR247 pKa = 11.84IYY249 pKa = 10.67FYY251 pKa = 11.26DD252 pKa = 3.51SVSNN256 pKa = 3.9
Molecular weight: 29.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1090 |
100 |
347 |
181.7 |
20.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.321 ± 0.524 | 2.11 ± 0.397 |
5.413 ± 0.697 | 4.404 ± 0.514 |
4.312 ± 0.584 | 5.321 ± 0.55 |
3.853 ± 0.71 | 4.954 ± 0.696 |
5.413 ± 1.192 | 7.706 ± 1.192 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.569 ± 0.558 | 5.229 ± 0.584 |
5.78 ± 0.62 | 4.22 ± 0.641 |
7.431 ± 1.176 | 8.44 ± 1.067 |
6.239 ± 0.951 | 6.055 ± 1.331 |
1.56 ± 0.201 | 3.67 ± 0.464 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
