
Cassava associated gemycircularvirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus cassa1
Average proteome isoelectric point is 7.46
Get precalculated fractions of proteins
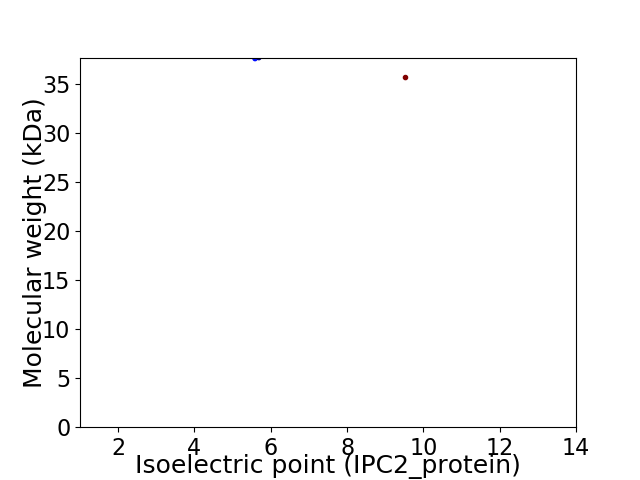
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I0AY94|I0AY94_9VIRU Capsid protein OS=Cassava associated gemycircularvirus 1 OX=1985374 PE=4 SV=1
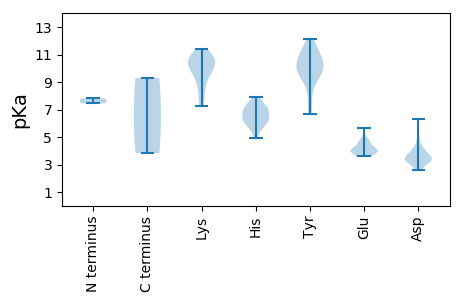
MM1 pKa = 7.81PFYY4 pKa = 11.01FNARR8 pKa = 11.84YY9 pKa = 9.8ALITYY14 pKa = 7.34AQCDD18 pKa = 3.64ALDD21 pKa = 3.81GFRR24 pKa = 11.84VMDD27 pKa = 4.02HH28 pKa = 6.63FSGLGAEE35 pKa = 4.67CIIARR40 pKa = 11.84EE41 pKa = 4.0IHH43 pKa = 5.9QDD45 pKa = 3.21GGVHH49 pKa = 5.75LHH51 pKa = 6.35CFIDD55 pKa = 4.11FGRR58 pKa = 11.84KK59 pKa = 7.81FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84TDD66 pKa = 2.64IFDD69 pKa = 3.14VDD71 pKa = 3.48GRR73 pKa = 11.84HH74 pKa = 6.37PNIEE78 pKa = 4.28PSWGTPWRR86 pKa = 11.84GYY88 pKa = 11.06DD89 pKa = 3.55YY90 pKa = 10.79AIKK93 pKa = 10.71DD94 pKa = 3.36GDD96 pKa = 4.29VICGGLEE103 pKa = 3.69RR104 pKa = 11.84PEE106 pKa = 3.96QPRR109 pKa = 11.84SEE111 pKa = 4.04RR112 pKa = 11.84VKK114 pKa = 10.93KK115 pKa = 10.61DD116 pKa = 2.57WDD118 pKa = 3.12QWAEE122 pKa = 3.65ITNARR127 pKa = 11.84DD128 pKa = 3.31RR129 pKa = 11.84DD130 pKa = 4.16HH131 pKa = 6.74FWEE134 pKa = 4.84LVHH137 pKa = 7.94HH138 pKa = 7.16LDD140 pKa = 4.88PKK142 pKa = 10.88AAACNYY148 pKa = 8.82GQLAKK153 pKa = 10.78YY154 pKa = 10.3ADD156 pKa = 3.4WRR158 pKa = 11.84FASVPPVYY166 pKa = 9.8EE167 pKa = 4.19SPGGISFIGGDD178 pKa = 3.39VDD180 pKa = 6.35GRR182 pKa = 11.84DD183 pKa = 3.28AWCEE187 pKa = 3.6QSHH190 pKa = 6.07IRR192 pKa = 11.84SGQPLPGRR200 pKa = 11.84CMSLVLYY207 pKa = 10.59GDD209 pKa = 3.49SRR211 pKa = 11.84TGKK214 pKa = 7.28TLWARR219 pKa = 11.84SLGTHH224 pKa = 7.44LYY226 pKa = 8.09TVGMVSGEE234 pKa = 4.09EE235 pKa = 3.83LKK237 pKa = 10.84KK238 pKa = 11.02ASNDD242 pKa = 3.4DD243 pKa = 3.12VKK245 pKa = 11.37YY246 pKa = 10.88AIFDD250 pKa = 4.73DD251 pKa = 3.41IRR253 pKa = 11.84GGIKK257 pKa = 9.97FFPAFKK263 pKa = 9.97EE264 pKa = 3.94WLGAQAYY271 pKa = 6.64VTVKK275 pKa = 10.24EE276 pKa = 4.66LYY278 pKa = 10.11RR279 pKa = 11.84EE280 pKa = 4.09PKK282 pKa = 8.72LVKK285 pKa = 9.08WGKK288 pKa = 8.36PSIWISNDD296 pKa = 3.14DD297 pKa = 3.81PRR299 pKa = 11.84LGMDD303 pKa = 3.57ASDD306 pKa = 4.25VSWLEE311 pKa = 3.81HH312 pKa = 4.93NCYY315 pKa = 9.61FVEE318 pKa = 4.32VSSPIFRR325 pKa = 11.84ANTEE329 pKa = 3.84
MM1 pKa = 7.81PFYY4 pKa = 11.01FNARR8 pKa = 11.84YY9 pKa = 9.8ALITYY14 pKa = 7.34AQCDD18 pKa = 3.64ALDD21 pKa = 3.81GFRR24 pKa = 11.84VMDD27 pKa = 4.02HH28 pKa = 6.63FSGLGAEE35 pKa = 4.67CIIARR40 pKa = 11.84EE41 pKa = 4.0IHH43 pKa = 5.9QDD45 pKa = 3.21GGVHH49 pKa = 5.75LHH51 pKa = 6.35CFIDD55 pKa = 4.11FGRR58 pKa = 11.84KK59 pKa = 7.81FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84TDD66 pKa = 2.64IFDD69 pKa = 3.14VDD71 pKa = 3.48GRR73 pKa = 11.84HH74 pKa = 6.37PNIEE78 pKa = 4.28PSWGTPWRR86 pKa = 11.84GYY88 pKa = 11.06DD89 pKa = 3.55YY90 pKa = 10.79AIKK93 pKa = 10.71DD94 pKa = 3.36GDD96 pKa = 4.29VICGGLEE103 pKa = 3.69RR104 pKa = 11.84PEE106 pKa = 3.96QPRR109 pKa = 11.84SEE111 pKa = 4.04RR112 pKa = 11.84VKK114 pKa = 10.93KK115 pKa = 10.61DD116 pKa = 2.57WDD118 pKa = 3.12QWAEE122 pKa = 3.65ITNARR127 pKa = 11.84DD128 pKa = 3.31RR129 pKa = 11.84DD130 pKa = 4.16HH131 pKa = 6.74FWEE134 pKa = 4.84LVHH137 pKa = 7.94HH138 pKa = 7.16LDD140 pKa = 4.88PKK142 pKa = 10.88AAACNYY148 pKa = 8.82GQLAKK153 pKa = 10.78YY154 pKa = 10.3ADD156 pKa = 3.4WRR158 pKa = 11.84FASVPPVYY166 pKa = 9.8EE167 pKa = 4.19SPGGISFIGGDD178 pKa = 3.39VDD180 pKa = 6.35GRR182 pKa = 11.84DD183 pKa = 3.28AWCEE187 pKa = 3.6QSHH190 pKa = 6.07IRR192 pKa = 11.84SGQPLPGRR200 pKa = 11.84CMSLVLYY207 pKa = 10.59GDD209 pKa = 3.49SRR211 pKa = 11.84TGKK214 pKa = 7.28TLWARR219 pKa = 11.84SLGTHH224 pKa = 7.44LYY226 pKa = 8.09TVGMVSGEE234 pKa = 4.09EE235 pKa = 3.83LKK237 pKa = 10.84KK238 pKa = 11.02ASNDD242 pKa = 3.4DD243 pKa = 3.12VKK245 pKa = 11.37YY246 pKa = 10.88AIFDD250 pKa = 4.73DD251 pKa = 3.41IRR253 pKa = 11.84GGIKK257 pKa = 9.97FFPAFKK263 pKa = 9.97EE264 pKa = 3.94WLGAQAYY271 pKa = 6.64VTVKK275 pKa = 10.24EE276 pKa = 4.66LYY278 pKa = 10.11RR279 pKa = 11.84EE280 pKa = 4.09PKK282 pKa = 8.72LVKK285 pKa = 9.08WGKK288 pKa = 8.36PSIWISNDD296 pKa = 3.14DD297 pKa = 3.81PRR299 pKa = 11.84LGMDD303 pKa = 3.57ASDD306 pKa = 4.25VSWLEE311 pKa = 3.81HH312 pKa = 4.93NCYY315 pKa = 9.61FVEE318 pKa = 4.32VSSPIFRR325 pKa = 11.84ANTEE329 pKa = 3.84
Molecular weight: 37.55 kDa
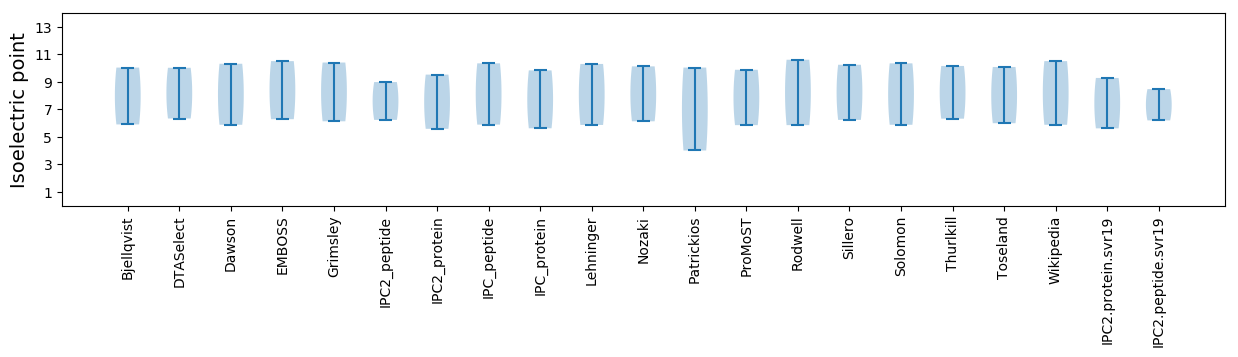
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0AY94|I0AY94_9VIRU Capsid protein OS=Cassava associated gemycircularvirus 1 OX=1985374 PE=4 SV=1
MM1 pKa = 7.45PRR3 pKa = 11.84ATRR6 pKa = 11.84TSDD9 pKa = 2.71RR10 pKa = 11.84LGRR13 pKa = 11.84GRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84STRR20 pKa = 11.84RR21 pKa = 11.84SSRR24 pKa = 11.84RR25 pKa = 11.84SVAKK29 pKa = 9.83RR30 pKa = 11.84PPYY33 pKa = 10.0RR34 pKa = 11.84KK35 pKa = 9.58YY36 pKa = 10.22LRR38 pKa = 11.84KK39 pKa = 8.54MSKK42 pKa = 10.01RR43 pKa = 11.84RR44 pKa = 11.84ILNITSTKK52 pKa = 10.55KK53 pKa = 9.82KK54 pKa = 10.02DD55 pKa = 3.3TMTGYY60 pKa = 9.34TNSTPANQTGGTVYY74 pKa = 9.77STDD77 pKa = 3.49PAIVTGGLNANQCNAFLWCATARR100 pKa = 11.84TNQVINPTQNTLVPGNKK117 pKa = 9.38FYY119 pKa = 9.93TACRR123 pKa = 11.84TDD125 pKa = 3.35SNPYY129 pKa = 9.07MVGLSEE135 pKa = 5.66KK136 pKa = 10.46IEE138 pKa = 4.03IQCNSGMPWQWRR150 pKa = 11.84RR151 pKa = 11.84ICWTQKK157 pKa = 10.34GGQGLPANSTSFNLSQQNAITGWNRR182 pKa = 11.84VMNQVAGNPGTGDD195 pKa = 3.28QYY197 pKa = 12.13NLFHH201 pKa = 7.06HH202 pKa = 7.12IFRR205 pKa = 11.84GQVGSDD211 pKa = 2.67WVDD214 pKa = 3.13VMTAPLDD221 pKa = 3.95PQHH224 pKa = 6.41ITVKK228 pKa = 9.45YY229 pKa = 10.02DD230 pKa = 3.29KK231 pKa = 10.99VVTLASGNEE240 pKa = 3.82EE241 pKa = 4.11GFIRR245 pKa = 11.84AYY247 pKa = 10.5RR248 pKa = 11.84RR249 pKa = 11.84WHH251 pKa = 5.98PMKK254 pKa = 10.01KK255 pKa = 7.35TLSYY259 pKa = 11.38ADD261 pKa = 3.68EE262 pKa = 4.69EE263 pKa = 4.68YY264 pKa = 11.53GNVEE268 pKa = 3.77NADD271 pKa = 3.64AFSQVGKK278 pKa = 10.64KK279 pKa = 10.81GMGDD283 pKa = 3.83YY284 pKa = 10.92YY285 pKa = 11.62VLDD288 pKa = 4.21LFRR291 pKa = 11.84ARR293 pKa = 11.84QGAQTNDD300 pKa = 3.66SISIRR305 pKa = 11.84PCATLYY311 pKa = 9.43WHH313 pKa = 7.09EE314 pKa = 4.25KK315 pKa = 9.3
MM1 pKa = 7.45PRR3 pKa = 11.84ATRR6 pKa = 11.84TSDD9 pKa = 2.71RR10 pKa = 11.84LGRR13 pKa = 11.84GRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84STRR20 pKa = 11.84RR21 pKa = 11.84SSRR24 pKa = 11.84RR25 pKa = 11.84SVAKK29 pKa = 9.83RR30 pKa = 11.84PPYY33 pKa = 10.0RR34 pKa = 11.84KK35 pKa = 9.58YY36 pKa = 10.22LRR38 pKa = 11.84KK39 pKa = 8.54MSKK42 pKa = 10.01RR43 pKa = 11.84RR44 pKa = 11.84ILNITSTKK52 pKa = 10.55KK53 pKa = 9.82KK54 pKa = 10.02DD55 pKa = 3.3TMTGYY60 pKa = 9.34TNSTPANQTGGTVYY74 pKa = 9.77STDD77 pKa = 3.49PAIVTGGLNANQCNAFLWCATARR100 pKa = 11.84TNQVINPTQNTLVPGNKK117 pKa = 9.38FYY119 pKa = 9.93TACRR123 pKa = 11.84TDD125 pKa = 3.35SNPYY129 pKa = 9.07MVGLSEE135 pKa = 5.66KK136 pKa = 10.46IEE138 pKa = 4.03IQCNSGMPWQWRR150 pKa = 11.84RR151 pKa = 11.84ICWTQKK157 pKa = 10.34GGQGLPANSTSFNLSQQNAITGWNRR182 pKa = 11.84VMNQVAGNPGTGDD195 pKa = 3.28QYY197 pKa = 12.13NLFHH201 pKa = 7.06HH202 pKa = 7.12IFRR205 pKa = 11.84GQVGSDD211 pKa = 2.67WVDD214 pKa = 3.13VMTAPLDD221 pKa = 3.95PQHH224 pKa = 6.41ITVKK228 pKa = 9.45YY229 pKa = 10.02DD230 pKa = 3.29KK231 pKa = 10.99VVTLASGNEE240 pKa = 3.82EE241 pKa = 4.11GFIRR245 pKa = 11.84AYY247 pKa = 10.5RR248 pKa = 11.84RR249 pKa = 11.84WHH251 pKa = 5.98PMKK254 pKa = 10.01KK255 pKa = 7.35TLSYY259 pKa = 11.38ADD261 pKa = 3.68EE262 pKa = 4.69EE263 pKa = 4.68YY264 pKa = 11.53GNVEE268 pKa = 3.77NADD271 pKa = 3.64AFSQVGKK278 pKa = 10.64KK279 pKa = 10.81GMGDD283 pKa = 3.83YY284 pKa = 10.92YY285 pKa = 11.62VLDD288 pKa = 4.21LFRR291 pKa = 11.84ARR293 pKa = 11.84QGAQTNDD300 pKa = 3.66SISIRR305 pKa = 11.84PCATLYY311 pKa = 9.43WHH313 pKa = 7.09EE314 pKa = 4.25KK315 pKa = 9.3
Molecular weight: 35.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
644 |
315 |
329 |
322.0 |
36.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.988 ± 0.227 | 2.174 ± 0.19 |
6.677 ± 1.576 | 4.037 ± 1.057 |
3.882 ± 0.947 | 8.696 ± 0.312 |
2.484 ± 0.633 | 4.969 ± 0.594 |
5.124 ± 0.192 | 5.59 ± 0.36 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.174 ± 0.482 | 4.814 ± 1.756 |
4.969 ± 0.146 | 3.882 ± 1.069 |
8.075 ± 0.575 | 6.366 ± 0.212 |
6.056 ± 2.224 | 5.59 ± 0.136 |
3.106 ± 0.399 | 4.348 ± 0.068 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
