
Lake Sarah-associated circular virus-20
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.78
Get precalculated fractions of proteins
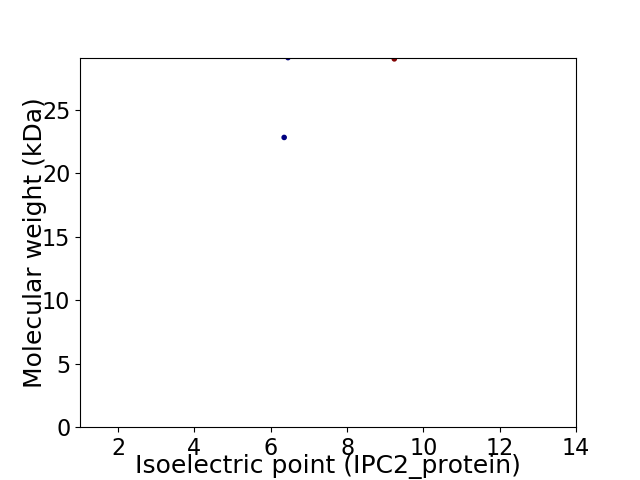
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A126G9P4|A0A126G9P4_9VIRU Coat protein OS=Lake Sarah-associated circular virus-20 OX=1685747 PE=4 SV=1
MM1 pKa = 7.81ADD3 pKa = 3.71KK4 pKa = 9.76STRR7 pKa = 11.84WAFTAYY13 pKa = 7.01EE14 pKa = 4.08TQYY17 pKa = 10.89PILDD21 pKa = 4.1ALHH24 pKa = 6.19KK25 pKa = 10.53QSSEE29 pKa = 4.21LIAEE33 pKa = 4.61IGWQDD38 pKa = 3.98EE39 pKa = 4.5ICPKK43 pKa = 9.34TGKK46 pKa = 8.8HH47 pKa = 5.3HH48 pKa = 5.88RR49 pKa = 11.84QGYY52 pKa = 8.0VRR54 pKa = 11.84TVRR57 pKa = 11.84QVRR60 pKa = 11.84FSQLRR65 pKa = 11.84EE66 pKa = 3.79IMPDD70 pKa = 2.64IHH72 pKa = 8.45LEE74 pKa = 3.96KK75 pKa = 10.6AKK77 pKa = 10.64NWEE80 pKa = 3.82ALINYY85 pKa = 6.83CQKK88 pKa = 10.79SATRR92 pKa = 11.84DD93 pKa = 3.23LSGAMVSAKK102 pKa = 9.91FEE104 pKa = 4.21RR105 pKa = 11.84PLRR108 pKa = 11.84LHH110 pKa = 6.85EE111 pKa = 4.06MLIDD115 pKa = 4.02VAHH118 pKa = 7.17HH119 pKa = 6.99IYY121 pKa = 10.51LSHH124 pKa = 7.65RR125 pKa = 11.84PGTKK129 pKa = 9.64HH130 pKa = 6.38AVCLDD135 pKa = 3.5RR136 pKa = 11.84QTDD139 pKa = 3.82RR140 pKa = 11.84QILLSYY146 pKa = 9.94LRR148 pKa = 11.84EE149 pKa = 3.8YY150 pKa = 10.96SAALVSAHH158 pKa = 6.93PEE160 pKa = 3.92YY161 pKa = 11.09AVVLVRR167 pKa = 11.84QDD169 pKa = 5.8ARR171 pKa = 11.84DD172 pKa = 3.16AWCNYY177 pKa = 8.85IEE179 pKa = 4.32VWMHH183 pKa = 5.9KK184 pKa = 10.43AEE186 pKa = 3.96QRR188 pKa = 11.84EE189 pKa = 4.25EE190 pKa = 4.26GDD192 pKa = 3.69EE193 pKa = 4.21VQQ195 pKa = 4.91
MM1 pKa = 7.81ADD3 pKa = 3.71KK4 pKa = 9.76STRR7 pKa = 11.84WAFTAYY13 pKa = 7.01EE14 pKa = 4.08TQYY17 pKa = 10.89PILDD21 pKa = 4.1ALHH24 pKa = 6.19KK25 pKa = 10.53QSSEE29 pKa = 4.21LIAEE33 pKa = 4.61IGWQDD38 pKa = 3.98EE39 pKa = 4.5ICPKK43 pKa = 9.34TGKK46 pKa = 8.8HH47 pKa = 5.3HH48 pKa = 5.88RR49 pKa = 11.84QGYY52 pKa = 8.0VRR54 pKa = 11.84TVRR57 pKa = 11.84QVRR60 pKa = 11.84FSQLRR65 pKa = 11.84EE66 pKa = 3.79IMPDD70 pKa = 2.64IHH72 pKa = 8.45LEE74 pKa = 3.96KK75 pKa = 10.6AKK77 pKa = 10.64NWEE80 pKa = 3.82ALINYY85 pKa = 6.83CQKK88 pKa = 10.79SATRR92 pKa = 11.84DD93 pKa = 3.23LSGAMVSAKK102 pKa = 9.91FEE104 pKa = 4.21RR105 pKa = 11.84PLRR108 pKa = 11.84LHH110 pKa = 6.85EE111 pKa = 4.06MLIDD115 pKa = 4.02VAHH118 pKa = 7.17HH119 pKa = 6.99IYY121 pKa = 10.51LSHH124 pKa = 7.65RR125 pKa = 11.84PGTKK129 pKa = 9.64HH130 pKa = 6.38AVCLDD135 pKa = 3.5RR136 pKa = 11.84QTDD139 pKa = 3.82RR140 pKa = 11.84QILLSYY146 pKa = 9.94LRR148 pKa = 11.84EE149 pKa = 3.8YY150 pKa = 10.96SAALVSAHH158 pKa = 6.93PEE160 pKa = 3.92YY161 pKa = 11.09AVVLVRR167 pKa = 11.84QDD169 pKa = 5.8ARR171 pKa = 11.84DD172 pKa = 3.16AWCNYY177 pKa = 8.85IEE179 pKa = 4.32VWMHH183 pKa = 5.9KK184 pKa = 10.43AEE186 pKa = 3.96QRR188 pKa = 11.84EE189 pKa = 4.25EE190 pKa = 4.26GDD192 pKa = 3.69EE193 pKa = 4.21VQQ195 pKa = 4.91
Molecular weight: 22.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G9P4|A0A126G9P4_9VIRU Coat protein OS=Lake Sarah-associated circular virus-20 OX=1685747 PE=4 SV=1
MM1 pKa = 7.29YY2 pKa = 10.63KK3 pKa = 9.65MVMAKK8 pKa = 10.02RR9 pKa = 11.84AFKK12 pKa = 10.15PRR14 pKa = 11.84RR15 pKa = 11.84QPRR18 pKa = 11.84RR19 pKa = 11.84VPRR22 pKa = 11.84APRR25 pKa = 11.84LRR27 pKa = 11.84RR28 pKa = 11.84ARR30 pKa = 11.84PTVEE34 pKa = 3.82FASAKK39 pKa = 7.42QTIALAQDD47 pKa = 3.04AMNQVFRR54 pKa = 11.84LDD56 pKa = 4.09NIALAQFDD64 pKa = 4.03RR65 pKa = 11.84MVNIAKK71 pKa = 9.84CYY73 pKa = 9.79QYY75 pKa = 11.41FRR77 pKa = 11.84MTKK80 pKa = 9.73VEE82 pKa = 4.76LKK84 pKa = 8.51FTPQMDD90 pKa = 3.71TFTDD94 pKa = 3.64SNLQSVPYY102 pKa = 10.05LYY104 pKa = 11.28YY105 pKa = 11.01LITKK109 pKa = 9.64GDD111 pKa = 3.8NLDD114 pKa = 3.8AGTFNKK120 pKa = 10.13LRR122 pKa = 11.84DD123 pKa = 3.74AGAKK127 pKa = 10.07GIRR130 pKa = 11.84FDD132 pKa = 4.79DD133 pKa = 4.17KK134 pKa = 11.26SINVNWRR141 pKa = 11.84PAVLRR146 pKa = 11.84GVIGEE151 pKa = 4.23DD152 pKa = 2.97TNAPVNPAFTAWAEE166 pKa = 4.28AKK168 pKa = 9.05TSPWLATSYY177 pKa = 11.0LPAAQSIVWSPSAVPHH193 pKa = 6.25KK194 pKa = 10.51GILYY198 pKa = 10.18GVEE201 pKa = 3.89QDD203 pKa = 3.72SSIDD207 pKa = 3.38TKK209 pKa = 11.42YY210 pKa = 11.04YY211 pKa = 11.07ALTLTAHH218 pKa = 6.34FQFKK222 pKa = 10.45KK223 pKa = 8.73PQTYY227 pKa = 8.15STTGVIDD234 pKa = 3.86PGATVKK240 pKa = 10.48QVIDD244 pKa = 3.49KK245 pKa = 10.73GDD247 pKa = 3.71VVEE250 pKa = 5.75LPLKK254 pKa = 10.01MLEE257 pKa = 4.05
MM1 pKa = 7.29YY2 pKa = 10.63KK3 pKa = 9.65MVMAKK8 pKa = 10.02RR9 pKa = 11.84AFKK12 pKa = 10.15PRR14 pKa = 11.84RR15 pKa = 11.84QPRR18 pKa = 11.84RR19 pKa = 11.84VPRR22 pKa = 11.84APRR25 pKa = 11.84LRR27 pKa = 11.84RR28 pKa = 11.84ARR30 pKa = 11.84PTVEE34 pKa = 3.82FASAKK39 pKa = 7.42QTIALAQDD47 pKa = 3.04AMNQVFRR54 pKa = 11.84LDD56 pKa = 4.09NIALAQFDD64 pKa = 4.03RR65 pKa = 11.84MVNIAKK71 pKa = 9.84CYY73 pKa = 9.79QYY75 pKa = 11.41FRR77 pKa = 11.84MTKK80 pKa = 9.73VEE82 pKa = 4.76LKK84 pKa = 8.51FTPQMDD90 pKa = 3.71TFTDD94 pKa = 3.64SNLQSVPYY102 pKa = 10.05LYY104 pKa = 11.28YY105 pKa = 11.01LITKK109 pKa = 9.64GDD111 pKa = 3.8NLDD114 pKa = 3.8AGTFNKK120 pKa = 10.13LRR122 pKa = 11.84DD123 pKa = 3.74AGAKK127 pKa = 10.07GIRR130 pKa = 11.84FDD132 pKa = 4.79DD133 pKa = 4.17KK134 pKa = 11.26SINVNWRR141 pKa = 11.84PAVLRR146 pKa = 11.84GVIGEE151 pKa = 4.23DD152 pKa = 2.97TNAPVNPAFTAWAEE166 pKa = 4.28AKK168 pKa = 9.05TSPWLATSYY177 pKa = 11.0LPAAQSIVWSPSAVPHH193 pKa = 6.25KK194 pKa = 10.51GILYY198 pKa = 10.18GVEE201 pKa = 3.89QDD203 pKa = 3.72SSIDD207 pKa = 3.38TKK209 pKa = 11.42YY210 pKa = 11.04YY211 pKa = 11.07ALTLTAHH218 pKa = 6.34FQFKK222 pKa = 10.45KK223 pKa = 8.73PQTYY227 pKa = 8.15STTGVIDD234 pKa = 3.86PGATVKK240 pKa = 10.48QVIDD244 pKa = 3.49KK245 pKa = 10.73GDD247 pKa = 3.71VVEE250 pKa = 5.75LPLKK254 pKa = 10.01MLEE257 pKa = 4.05
Molecular weight: 29.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
452 |
195 |
257 |
226.0 |
25.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.398 ± 0.427 | 1.106 ± 0.617 |
5.973 ± 0.217 | 5.088 ± 2.034 |
3.319 ± 1.162 | 3.761 ± 0.446 |
2.876 ± 1.804 | 5.088 ± 0.361 |
6.416 ± 0.84 | 7.965 ± 0.492 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.876 ± 0.204 | 2.876 ± 0.873 |
5.088 ± 1.313 | 5.531 ± 0.406 |
7.301 ± 0.59 | 5.088 ± 0.361 |
5.973 ± 1.221 | 6.858 ± 0.46 |
1.991 ± 0.374 | 4.425 ± 0.124 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
