
Lewinella marina
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Saprospiria; Saprospirales; Lewinellaceae; Lewinella
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
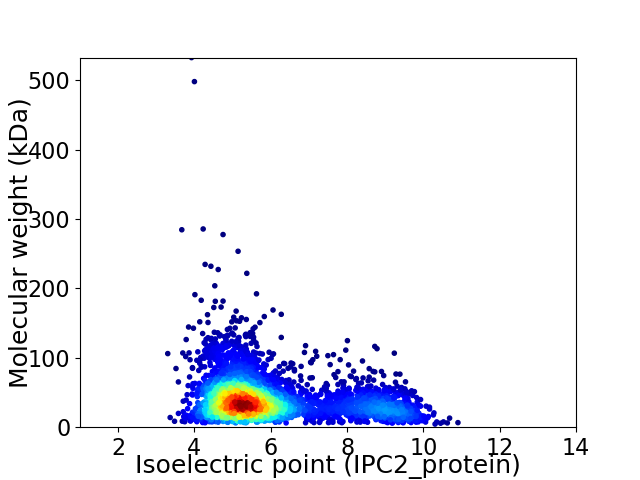
Virtual 2D-PAGE plot for 3525 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2G0CBM1|A0A2G0CBM1_9BACT Phosphoribosylformylglycinamidine synthase subunit PurQ OS=Lewinella marina OX=438751 GN=purQ PE=3 SV=1
MM1 pKa = 7.38KK2 pKa = 9.41PDD4 pKa = 3.44KK5 pKa = 8.84PTLVGSSSRR14 pKa = 11.84RR15 pKa = 11.84PLPAAALRR23 pKa = 11.84MLMNYY28 pKa = 8.03TFMPDD33 pKa = 3.03RR34 pKa = 11.84PGIRR38 pKa = 11.84ALFLLLFAIFGVTGLRR54 pKa = 11.84AQLALTVTKK63 pKa = 10.45TDD65 pKa = 3.41ISCLGEE71 pKa = 3.82RR72 pKa = 11.84DD73 pKa = 3.62GVITVSARR81 pKa = 11.84GGSPDD86 pKa = 3.43YY87 pKa = 10.93DD88 pKa = 3.73FSIVRR93 pKa = 11.84TRR95 pKa = 11.84PATPPVTGSNSTNSDD110 pKa = 2.58GVAINFTNLNSGEE123 pKa = 4.19YY124 pKa = 9.3TVTVTDD130 pKa = 3.74AAGNTATDD138 pKa = 3.38QVEE141 pKa = 4.05ILEE144 pKa = 4.42PNITTAIQVVQEE156 pKa = 4.09IQCFDD161 pKa = 3.63DD162 pKa = 3.58TNGILEE168 pKa = 4.19AVVFDD173 pKa = 4.03GSNPISDD180 pKa = 3.52YY181 pKa = 11.57SNYY184 pKa = 7.8TFRR187 pKa = 11.84WSTGAITRR195 pKa = 11.84RR196 pKa = 11.84ISALGPDD203 pKa = 3.86QYY205 pKa = 11.75SVTVTNTTTGCASSDD220 pKa = 3.85SEE222 pKa = 4.45TLRR225 pKa = 11.84SPSRR229 pKa = 11.84LVANSSQLRR238 pKa = 11.84SDD240 pKa = 3.66AATCSGVSDD249 pKa = 3.69GTVNVPITGGTKK261 pKa = 8.49NTNGNYY267 pKa = 9.93RR268 pKa = 11.84INWSDD273 pKa = 3.23GTEE276 pKa = 4.0TLDD279 pKa = 3.91NKK281 pKa = 10.13IARR284 pKa = 11.84SDD286 pKa = 4.15LEE288 pKa = 4.47PGSYY292 pKa = 9.06TVTVTDD298 pKa = 4.0ARR300 pKa = 11.84SCEE303 pKa = 4.21TTTSFTVAAEE313 pKa = 4.01KK314 pKa = 10.15TLLLTAATTDD324 pKa = 3.51ISCFGDD330 pKa = 3.3ANGSISVTAGFEE342 pKa = 4.24GATPDD347 pKa = 3.56YY348 pKa = 10.7PFEE351 pKa = 4.24TRR353 pKa = 11.84LLAEE357 pKa = 5.6DD358 pKa = 3.55GTEE361 pKa = 3.77LVGYY365 pKa = 7.38QTVANQGRR373 pKa = 11.84TPRR376 pKa = 11.84VFDD379 pKa = 3.42NLGPGNYY386 pKa = 9.67LVVLRR391 pKa = 11.84DD392 pKa = 3.64QDD394 pKa = 4.6SEE396 pKa = 4.76GCTVTDD402 pKa = 3.36TVTIIEE408 pKa = 4.34PALLEE413 pKa = 4.33IDD415 pKa = 3.83TVEE418 pKa = 4.31TTDD421 pKa = 4.49FGCPDD426 pKa = 3.62EE427 pKa = 5.8FGTATINFTGGTAPYY442 pKa = 9.67TFRR445 pKa = 11.84FKK447 pKa = 10.99NDD449 pKa = 3.65SLPHH453 pKa = 5.77PTDD456 pKa = 2.68TTMRR460 pKa = 11.84FDD462 pKa = 4.46TIVADD467 pKa = 4.42TNFLDD472 pKa = 4.76QLQPDD477 pKa = 4.06TNYY480 pKa = 10.41VAFVTDD486 pKa = 4.18ANGCIDD492 pKa = 3.57SLRR495 pKa = 11.84FKK497 pKa = 10.59IFSPPRR503 pKa = 11.84ASIQPIATDD512 pKa = 3.73SVSCPSSSDD521 pKa = 3.2GQLFTIVTPPQGEE534 pKa = 4.7TVTEE538 pKa = 4.22TAWYY542 pKa = 9.66RR543 pKa = 11.84INNDD547 pKa = 2.84GTIGEE552 pKa = 4.27QVDD555 pKa = 2.87IGTRR559 pKa = 11.84TTADD563 pKa = 3.51LSVGFYY569 pKa = 10.7LFEE572 pKa = 4.43ATLSNACVSQAIGEE586 pKa = 4.4VASPGLVEE594 pKa = 5.66LDD596 pKa = 4.1SFDD599 pKa = 4.13LQPPVCLGEE608 pKa = 4.42SNGSIFVYY616 pKa = 8.73PTGGTPPYY624 pKa = 10.2RR625 pKa = 11.84YY626 pKa = 9.25DD627 pKa = 3.02WSVAGASTSANFITNLSAGTYY648 pKa = 10.16SVTITDD654 pKa = 4.9ANNCQPPFSTTFVLEE669 pKa = 4.32DD670 pKa = 3.44PVGITGTFSDD680 pKa = 4.63LQGVSCPDD688 pKa = 3.17EE689 pKa = 4.29STADD693 pKa = 3.48GAATFDD699 pKa = 4.09ALFSDD704 pKa = 4.91GSTGTFDD711 pKa = 5.35FYY713 pKa = 10.72WSSGDD718 pKa = 3.69TITGQSSATVTGLTRR733 pKa = 11.84GPISVTVTDD742 pKa = 4.72GLCPQVFTDD751 pKa = 5.09TISSPEE757 pKa = 3.85DD758 pKa = 3.31FEE760 pKa = 5.16IAFATEE766 pKa = 4.18DD767 pKa = 3.88VNCFGQSDD775 pKa = 4.39GSVTAQVTGGTPGYY789 pKa = 8.45TYY791 pKa = 11.29NWIDD795 pKa = 3.66RR796 pKa = 11.84PEE798 pKa = 4.23TGPAITGVPAGSYY811 pKa = 10.2QLLITDD817 pKa = 4.83ARR819 pKa = 11.84GCQPDD824 pKa = 3.51TAAAVVNQPDD834 pKa = 4.6PLTLSINPTLTTPTVTCAGDD854 pKa = 3.64SNGVVAVFVSSTNNNPLANNPYY876 pKa = 9.01RR877 pKa = 11.84WSANVADD884 pKa = 4.76NDD886 pKa = 4.44DD887 pKa = 3.88NVAGNLAPGTYY898 pKa = 10.21AVTVTDD904 pKa = 4.64VEE906 pKa = 4.76GCQDD910 pKa = 3.36SLQYY914 pKa = 10.44TIIEE918 pKa = 4.15PQPITFAVQDD928 pKa = 4.34IEE930 pKa = 4.39PPLCFGEE937 pKa = 4.35MTTVRR942 pKa = 11.84LDD944 pKa = 3.41TAFGGQSSGFDD955 pKa = 3.31DD956 pKa = 4.93FTFTLNNDD964 pKa = 3.57GFLLPADD971 pKa = 3.87QPGSTFAGEE980 pKa = 4.3VVVTVFDD987 pKa = 4.27SVGCRR992 pKa = 11.84SDD994 pKa = 3.09LTFNVEE1000 pKa = 3.94QPEE1003 pKa = 4.41EE1004 pKa = 4.01IVIEE1008 pKa = 4.16LPEE1011 pKa = 4.75RR1012 pKa = 11.84LLVDD1016 pKa = 4.85LGDD1019 pKa = 3.89TLEE1022 pKa = 4.05QLTPVVTPMSSNYY1035 pKa = 10.29SYY1037 pKa = 11.37LWTPAEE1043 pKa = 4.23FLSADD1048 pKa = 3.5TVANPFVTPLQTRR1061 pKa = 11.84IYY1063 pKa = 8.69TLTVTDD1069 pKa = 3.68QNGCEE1074 pKa = 4.17AIEE1077 pKa = 5.05DD1078 pKa = 3.68ILVEE1082 pKa = 3.81VDD1084 pKa = 3.17ANRR1087 pKa = 11.84NVYY1090 pKa = 9.97LPNAFSPNQDD1100 pKa = 2.51GRR1102 pKa = 11.84NEE1104 pKa = 3.82EE1105 pKa = 3.92FRR1107 pKa = 11.84IYY1109 pKa = 10.7ACQGVVGVPRR1119 pKa = 11.84VNIFNRR1125 pKa = 11.84WGGLVYY1131 pKa = 10.32EE1132 pKa = 4.82ATDD1135 pKa = 3.53IPANCLDD1142 pKa = 4.64GTLLWDD1148 pKa = 3.53GRR1150 pKa = 11.84TPSGQLADD1158 pKa = 4.0LGVYY1162 pKa = 10.16VYY1164 pKa = 9.5TVEE1167 pKa = 3.92VAFIDD1172 pKa = 4.59GVTLLYY1178 pKa = 10.62RR1179 pKa = 11.84GDD1181 pKa = 3.4IAVIRR1186 pKa = 4.25
MM1 pKa = 7.38KK2 pKa = 9.41PDD4 pKa = 3.44KK5 pKa = 8.84PTLVGSSSRR14 pKa = 11.84RR15 pKa = 11.84PLPAAALRR23 pKa = 11.84MLMNYY28 pKa = 8.03TFMPDD33 pKa = 3.03RR34 pKa = 11.84PGIRR38 pKa = 11.84ALFLLLFAIFGVTGLRR54 pKa = 11.84AQLALTVTKK63 pKa = 10.45TDD65 pKa = 3.41ISCLGEE71 pKa = 3.82RR72 pKa = 11.84DD73 pKa = 3.62GVITVSARR81 pKa = 11.84GGSPDD86 pKa = 3.43YY87 pKa = 10.93DD88 pKa = 3.73FSIVRR93 pKa = 11.84TRR95 pKa = 11.84PATPPVTGSNSTNSDD110 pKa = 2.58GVAINFTNLNSGEE123 pKa = 4.19YY124 pKa = 9.3TVTVTDD130 pKa = 3.74AAGNTATDD138 pKa = 3.38QVEE141 pKa = 4.05ILEE144 pKa = 4.42PNITTAIQVVQEE156 pKa = 4.09IQCFDD161 pKa = 3.63DD162 pKa = 3.58TNGILEE168 pKa = 4.19AVVFDD173 pKa = 4.03GSNPISDD180 pKa = 3.52YY181 pKa = 11.57SNYY184 pKa = 7.8TFRR187 pKa = 11.84WSTGAITRR195 pKa = 11.84RR196 pKa = 11.84ISALGPDD203 pKa = 3.86QYY205 pKa = 11.75SVTVTNTTTGCASSDD220 pKa = 3.85SEE222 pKa = 4.45TLRR225 pKa = 11.84SPSRR229 pKa = 11.84LVANSSQLRR238 pKa = 11.84SDD240 pKa = 3.66AATCSGVSDD249 pKa = 3.69GTVNVPITGGTKK261 pKa = 8.49NTNGNYY267 pKa = 9.93RR268 pKa = 11.84INWSDD273 pKa = 3.23GTEE276 pKa = 4.0TLDD279 pKa = 3.91NKK281 pKa = 10.13IARR284 pKa = 11.84SDD286 pKa = 4.15LEE288 pKa = 4.47PGSYY292 pKa = 9.06TVTVTDD298 pKa = 4.0ARR300 pKa = 11.84SCEE303 pKa = 4.21TTTSFTVAAEE313 pKa = 4.01KK314 pKa = 10.15TLLLTAATTDD324 pKa = 3.51ISCFGDD330 pKa = 3.3ANGSISVTAGFEE342 pKa = 4.24GATPDD347 pKa = 3.56YY348 pKa = 10.7PFEE351 pKa = 4.24TRR353 pKa = 11.84LLAEE357 pKa = 5.6DD358 pKa = 3.55GTEE361 pKa = 3.77LVGYY365 pKa = 7.38QTVANQGRR373 pKa = 11.84TPRR376 pKa = 11.84VFDD379 pKa = 3.42NLGPGNYY386 pKa = 9.67LVVLRR391 pKa = 11.84DD392 pKa = 3.64QDD394 pKa = 4.6SEE396 pKa = 4.76GCTVTDD402 pKa = 3.36TVTIIEE408 pKa = 4.34PALLEE413 pKa = 4.33IDD415 pKa = 3.83TVEE418 pKa = 4.31TTDD421 pKa = 4.49FGCPDD426 pKa = 3.62EE427 pKa = 5.8FGTATINFTGGTAPYY442 pKa = 9.67TFRR445 pKa = 11.84FKK447 pKa = 10.99NDD449 pKa = 3.65SLPHH453 pKa = 5.77PTDD456 pKa = 2.68TTMRR460 pKa = 11.84FDD462 pKa = 4.46TIVADD467 pKa = 4.42TNFLDD472 pKa = 4.76QLQPDD477 pKa = 4.06TNYY480 pKa = 10.41VAFVTDD486 pKa = 4.18ANGCIDD492 pKa = 3.57SLRR495 pKa = 11.84FKK497 pKa = 10.59IFSPPRR503 pKa = 11.84ASIQPIATDD512 pKa = 3.73SVSCPSSSDD521 pKa = 3.2GQLFTIVTPPQGEE534 pKa = 4.7TVTEE538 pKa = 4.22TAWYY542 pKa = 9.66RR543 pKa = 11.84INNDD547 pKa = 2.84GTIGEE552 pKa = 4.27QVDD555 pKa = 2.87IGTRR559 pKa = 11.84TTADD563 pKa = 3.51LSVGFYY569 pKa = 10.7LFEE572 pKa = 4.43ATLSNACVSQAIGEE586 pKa = 4.4VASPGLVEE594 pKa = 5.66LDD596 pKa = 4.1SFDD599 pKa = 4.13LQPPVCLGEE608 pKa = 4.42SNGSIFVYY616 pKa = 8.73PTGGTPPYY624 pKa = 10.2RR625 pKa = 11.84YY626 pKa = 9.25DD627 pKa = 3.02WSVAGASTSANFITNLSAGTYY648 pKa = 10.16SVTITDD654 pKa = 4.9ANNCQPPFSTTFVLEE669 pKa = 4.32DD670 pKa = 3.44PVGITGTFSDD680 pKa = 4.63LQGVSCPDD688 pKa = 3.17EE689 pKa = 4.29STADD693 pKa = 3.48GAATFDD699 pKa = 4.09ALFSDD704 pKa = 4.91GSTGTFDD711 pKa = 5.35FYY713 pKa = 10.72WSSGDD718 pKa = 3.69TITGQSSATVTGLTRR733 pKa = 11.84GPISVTVTDD742 pKa = 4.72GLCPQVFTDD751 pKa = 5.09TISSPEE757 pKa = 3.85DD758 pKa = 3.31FEE760 pKa = 5.16IAFATEE766 pKa = 4.18DD767 pKa = 3.88VNCFGQSDD775 pKa = 4.39GSVTAQVTGGTPGYY789 pKa = 8.45TYY791 pKa = 11.29NWIDD795 pKa = 3.66RR796 pKa = 11.84PEE798 pKa = 4.23TGPAITGVPAGSYY811 pKa = 10.2QLLITDD817 pKa = 4.83ARR819 pKa = 11.84GCQPDD824 pKa = 3.51TAAAVVNQPDD834 pKa = 4.6PLTLSINPTLTTPTVTCAGDD854 pKa = 3.64SNGVVAVFVSSTNNNPLANNPYY876 pKa = 9.01RR877 pKa = 11.84WSANVADD884 pKa = 4.76NDD886 pKa = 4.44DD887 pKa = 3.88NVAGNLAPGTYY898 pKa = 10.21AVTVTDD904 pKa = 4.64VEE906 pKa = 4.76GCQDD910 pKa = 3.36SLQYY914 pKa = 10.44TIIEE918 pKa = 4.15PQPITFAVQDD928 pKa = 4.34IEE930 pKa = 4.39PPLCFGEE937 pKa = 4.35MTTVRR942 pKa = 11.84LDD944 pKa = 3.41TAFGGQSSGFDD955 pKa = 3.31DD956 pKa = 4.93FTFTLNNDD964 pKa = 3.57GFLLPADD971 pKa = 3.87QPGSTFAGEE980 pKa = 4.3VVVTVFDD987 pKa = 4.27SVGCRR992 pKa = 11.84SDD994 pKa = 3.09LTFNVEE1000 pKa = 3.94QPEE1003 pKa = 4.41EE1004 pKa = 4.01IVIEE1008 pKa = 4.16LPEE1011 pKa = 4.75RR1012 pKa = 11.84LLVDD1016 pKa = 4.85LGDD1019 pKa = 3.89TLEE1022 pKa = 4.05QLTPVVTPMSSNYY1035 pKa = 10.29SYY1037 pKa = 11.37LWTPAEE1043 pKa = 4.23FLSADD1048 pKa = 3.5TVANPFVTPLQTRR1061 pKa = 11.84IYY1063 pKa = 8.69TLTVTDD1069 pKa = 3.68QNGCEE1074 pKa = 4.17AIEE1077 pKa = 5.05DD1078 pKa = 3.68ILVEE1082 pKa = 3.81VDD1084 pKa = 3.17ANRR1087 pKa = 11.84NVYY1090 pKa = 9.97LPNAFSPNQDD1100 pKa = 2.51GRR1102 pKa = 11.84NEE1104 pKa = 3.82EE1105 pKa = 3.92FRR1107 pKa = 11.84IYY1109 pKa = 10.7ACQGVVGVPRR1119 pKa = 11.84VNIFNRR1125 pKa = 11.84WGGLVYY1131 pKa = 10.32EE1132 pKa = 4.82ATDD1135 pKa = 3.53IPANCLDD1142 pKa = 4.64GTLLWDD1148 pKa = 3.53GRR1150 pKa = 11.84TPSGQLADD1158 pKa = 4.0LGVYY1162 pKa = 10.16VYY1164 pKa = 9.5TVEE1167 pKa = 3.92VAFIDD1172 pKa = 4.59GVTLLYY1178 pKa = 10.62RR1179 pKa = 11.84GDD1181 pKa = 3.4IAVIRR1186 pKa = 4.25
Molecular weight: 126.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2G0CDI0|A0A2G0CDI0_9BACT Carrier domain-containing protein OS=Lewinella marina OX=438751 GN=CGL56_12685 PE=4 SV=1
MM1 pKa = 7.47SSAGRR6 pKa = 11.84KK7 pKa = 8.54GGFRR11 pKa = 11.84RR12 pKa = 11.84FYY14 pKa = 8.4WLSRR18 pKa = 11.84VFPGQEE24 pKa = 3.84GQQFGAYY31 pKa = 8.71RR32 pKa = 11.84RR33 pKa = 11.84LVRR36 pKa = 11.84EE37 pKa = 3.76NNIRR41 pKa = 11.84CPQGARR47 pKa = 11.84RR48 pKa = 11.84VDD50 pKa = 3.59RR51 pKa = 11.84HH52 pKa = 5.63PMSVEE57 pKa = 3.14PRR59 pKa = 11.84EE60 pKa = 4.19FVEE63 pKa = 5.53KK64 pKa = 10.81NHH66 pKa = 6.14SPCNKK71 pKa = 9.4APRR74 pKa = 11.84LLRR77 pKa = 11.84VPDD80 pKa = 3.54KK81 pKa = 10.48RR82 pKa = 11.84TGVQSFSSLVEE93 pKa = 3.96SVRR96 pKa = 11.84DD97 pKa = 3.38RR98 pKa = 11.84LYY100 pKa = 10.81RR101 pKa = 11.84LAVRR105 pKa = 11.84MTGDD109 pKa = 3.03GSEE112 pKa = 4.61AEE114 pKa = 4.58DD115 pKa = 3.66VVQEE119 pKa = 3.99VLISGWQRR127 pKa = 11.84KK128 pKa = 7.38EE129 pKa = 4.27EE130 pKa = 3.97IVQLEE135 pKa = 4.33NPPAWLMRR143 pKa = 11.84MTHH146 pKa = 5.38NRR148 pKa = 11.84AIDD151 pKa = 3.38RR152 pKa = 11.84LRR154 pKa = 11.84SRR156 pKa = 11.84KK157 pKa = 9.52ARR159 pKa = 11.84TGYY162 pKa = 9.82EE163 pKa = 3.74RR164 pKa = 11.84AAAPADD170 pKa = 3.81YY171 pKa = 10.71HH172 pKa = 6.76GLTPLRR178 pKa = 11.84LLEE181 pKa = 4.32TRR183 pKa = 11.84DD184 pKa = 3.71TLDD187 pKa = 4.28LIHH190 pKa = 6.87RR191 pKa = 11.84QMARR195 pKa = 11.84LSPEE199 pKa = 3.69QRR201 pKa = 11.84SVLQLRR207 pKa = 11.84EE208 pKa = 4.13IEE210 pKa = 4.13GMSYY214 pKa = 10.74RR215 pKa = 11.84EE216 pKa = 3.77IAEE219 pKa = 4.02ATGLSTEE226 pKa = 4.04QVKK229 pKa = 10.46VYY231 pKa = 8.63LHH233 pKa = 6.48RR234 pKa = 11.84GRR236 pKa = 11.84RR237 pKa = 11.84QLRR240 pKa = 11.84QWLLQEE246 pKa = 4.87KK247 pKa = 9.94IVEE250 pKa = 4.21RR251 pKa = 4.29
MM1 pKa = 7.47SSAGRR6 pKa = 11.84KK7 pKa = 8.54GGFRR11 pKa = 11.84RR12 pKa = 11.84FYY14 pKa = 8.4WLSRR18 pKa = 11.84VFPGQEE24 pKa = 3.84GQQFGAYY31 pKa = 8.71RR32 pKa = 11.84RR33 pKa = 11.84LVRR36 pKa = 11.84EE37 pKa = 3.76NNIRR41 pKa = 11.84CPQGARR47 pKa = 11.84RR48 pKa = 11.84VDD50 pKa = 3.59RR51 pKa = 11.84HH52 pKa = 5.63PMSVEE57 pKa = 3.14PRR59 pKa = 11.84EE60 pKa = 4.19FVEE63 pKa = 5.53KK64 pKa = 10.81NHH66 pKa = 6.14SPCNKK71 pKa = 9.4APRR74 pKa = 11.84LLRR77 pKa = 11.84VPDD80 pKa = 3.54KK81 pKa = 10.48RR82 pKa = 11.84TGVQSFSSLVEE93 pKa = 3.96SVRR96 pKa = 11.84DD97 pKa = 3.38RR98 pKa = 11.84LYY100 pKa = 10.81RR101 pKa = 11.84LAVRR105 pKa = 11.84MTGDD109 pKa = 3.03GSEE112 pKa = 4.61AEE114 pKa = 4.58DD115 pKa = 3.66VVQEE119 pKa = 3.99VLISGWQRR127 pKa = 11.84KK128 pKa = 7.38EE129 pKa = 4.27EE130 pKa = 3.97IVQLEE135 pKa = 4.33NPPAWLMRR143 pKa = 11.84MTHH146 pKa = 5.38NRR148 pKa = 11.84AIDD151 pKa = 3.38RR152 pKa = 11.84LRR154 pKa = 11.84SRR156 pKa = 11.84KK157 pKa = 9.52ARR159 pKa = 11.84TGYY162 pKa = 9.82EE163 pKa = 3.74RR164 pKa = 11.84AAAPADD170 pKa = 3.81YY171 pKa = 10.71HH172 pKa = 6.76GLTPLRR178 pKa = 11.84LLEE181 pKa = 4.32TRR183 pKa = 11.84DD184 pKa = 3.71TLDD187 pKa = 4.28LIHH190 pKa = 6.87RR191 pKa = 11.84QMARR195 pKa = 11.84LSPEE199 pKa = 3.69QRR201 pKa = 11.84SVLQLRR207 pKa = 11.84EE208 pKa = 4.13IEE210 pKa = 4.13GMSYY214 pKa = 10.74RR215 pKa = 11.84EE216 pKa = 3.77IAEE219 pKa = 4.02ATGLSTEE226 pKa = 4.04QVKK229 pKa = 10.46VYY231 pKa = 8.63LHH233 pKa = 6.48RR234 pKa = 11.84GRR236 pKa = 11.84RR237 pKa = 11.84QLRR240 pKa = 11.84QWLLQEE246 pKa = 4.87KK247 pKa = 9.94IVEE250 pKa = 4.21RR251 pKa = 4.29
Molecular weight: 29.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1348891 |
38 |
5003 |
382.7 |
42.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.195 ± 0.046 | 0.763 ± 0.014 |
6.094 ± 0.036 | 6.371 ± 0.04 |
4.237 ± 0.025 | 8.088 ± 0.037 |
2.084 ± 0.021 | 4.973 ± 0.029 |
3.195 ± 0.033 | 10.409 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.947 ± 0.018 | 3.787 ± 0.034 |
5.012 ± 0.029 | 3.564 ± 0.023 |
6.797 ± 0.043 | 5.522 ± 0.028 |
5.907 ± 0.038 | 6.939 ± 0.028 |
1.313 ± 0.016 | 3.801 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
