
Cauliflower mosaic virus (strain CM-1841) (CaMV)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Caulimovirus; Cauliflower mosaic virus
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
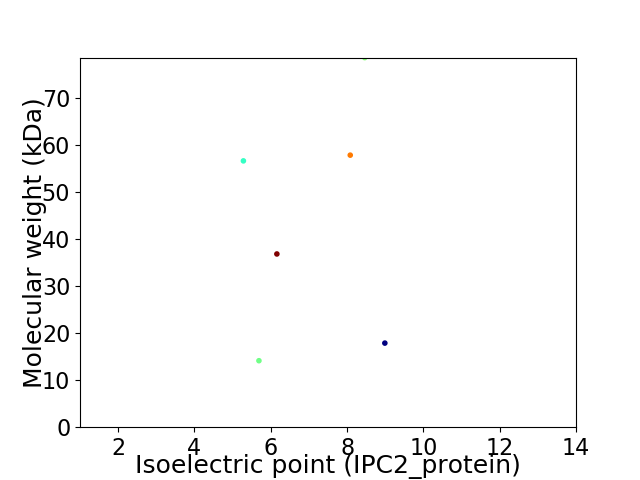
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
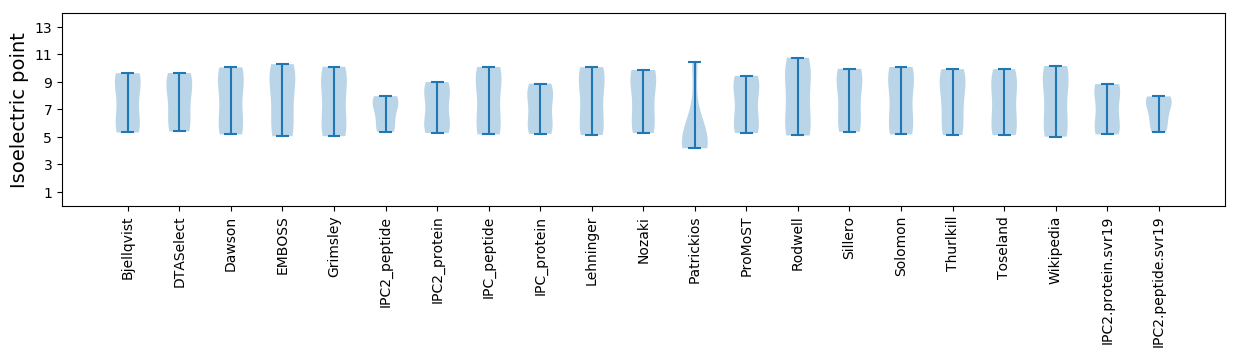
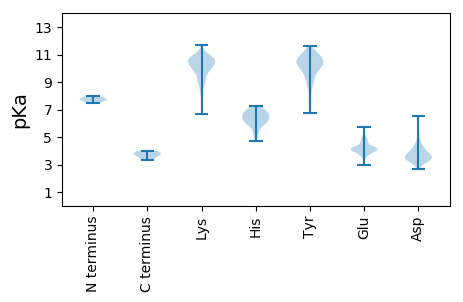
Protein with the lowest isoelectric point:
>sp|P03546|MVP_CAMVC Movement protein OS=Cauliflower mosaic virus (strain CM-1841) OX=10644 GN=ORF I PE=1 SV=1
MM1 pKa = 7.73AEE3 pKa = 4.5SILDD7 pKa = 3.48RR8 pKa = 11.84TINRR12 pKa = 11.84FWYY15 pKa = 10.5NLGEE19 pKa = 4.56DD20 pKa = 4.62CLSEE24 pKa = 4.25SQFDD28 pKa = 3.63LMIRR32 pKa = 11.84LMEE35 pKa = 4.36EE36 pKa = 4.14SLDD39 pKa = 3.54GDD41 pKa = 3.95QIIDD45 pKa = 3.92LTSLPSDD52 pKa = 3.49NLQVEE57 pKa = 4.7QVMTTTDD64 pKa = 3.35DD65 pKa = 4.2SISEE69 pKa = 4.02EE70 pKa = 4.57SEE72 pKa = 3.64FLLAIGEE79 pKa = 4.16ISEE82 pKa = 4.84DD83 pKa = 3.61EE84 pKa = 4.21SDD86 pKa = 3.66SGEE89 pKa = 4.04EE90 pKa = 4.03PEE92 pKa = 4.39FEE94 pKa = 4.26QVRR97 pKa = 11.84MDD99 pKa = 3.4RR100 pKa = 11.84TGGTEE105 pKa = 3.59IPKK108 pKa = 10.47EE109 pKa = 3.95EE110 pKa = 4.82DD111 pKa = 3.31GEE113 pKa = 4.73GPSRR117 pKa = 11.84YY118 pKa = 9.29NEE120 pKa = 4.24RR121 pKa = 11.84KK122 pKa = 9.81RR123 pKa = 11.84KK124 pKa = 7.47TPEE127 pKa = 3.29DD128 pKa = 4.14RR129 pKa = 11.84YY130 pKa = 10.74FPTQPKK136 pKa = 8.96TIPGQKK142 pKa = 6.7QTSMGMLNIDD152 pKa = 3.45CQINRR157 pKa = 11.84RR158 pKa = 11.84TLIDD162 pKa = 3.36DD163 pKa = 4.03WAAEE167 pKa = 3.64IGLIVKK173 pKa = 7.4TNRR176 pKa = 11.84EE177 pKa = 4.21DD178 pKa = 3.71YY179 pKa = 10.97LDD181 pKa = 4.59PEE183 pKa = 4.86TILLLMEE190 pKa = 5.35HH191 pKa = 6.19KK192 pKa = 9.65TSGIAKK198 pKa = 8.99EE199 pKa = 4.39LIRR202 pKa = 11.84NTRR205 pKa = 11.84WNRR208 pKa = 11.84TTGDD212 pKa = 3.23IIEE215 pKa = 4.15QVINAMYY222 pKa = 10.9TMFLGLNYY230 pKa = 10.01SDD232 pKa = 4.16NKK234 pKa = 9.76VAEE237 pKa = 4.95KK238 pKa = 9.96IDD240 pKa = 3.71EE241 pKa = 4.11QEE243 pKa = 3.87KK244 pKa = 10.81AKK246 pKa = 10.57IRR248 pKa = 11.84MTKK251 pKa = 10.09LQLFDD256 pKa = 2.96ICYY259 pKa = 10.29LEE261 pKa = 5.62EE262 pKa = 4.13FTCDD266 pKa = 3.39YY267 pKa = 9.76EE268 pKa = 5.13KK269 pKa = 11.56NMYY272 pKa = 7.76KK273 pKa = 9.52TEE275 pKa = 3.96MADD278 pKa = 3.35FPGYY282 pKa = 10.03INQYY286 pKa = 9.44LSKK289 pKa = 10.49IPIIGEE295 pKa = 3.82KK296 pKa = 10.07ALTRR300 pKa = 11.84FRR302 pKa = 11.84HH303 pKa = 5.1EE304 pKa = 4.41ANGTSIYY311 pKa = 10.98SLGFAAKK318 pKa = 9.37IVKK321 pKa = 10.16EE322 pKa = 4.25EE323 pKa = 3.98LSKK326 pKa = 10.93ICDD329 pKa = 3.44LSKK332 pKa = 10.64KK333 pKa = 8.32QKK335 pKa = 10.13KK336 pKa = 9.23LKK338 pKa = 10.43KK339 pKa = 9.92FNKK342 pKa = 8.29KK343 pKa = 9.46CCSIGEE349 pKa = 4.05ASVEE353 pKa = 3.97YY354 pKa = 10.54GGKK357 pKa = 8.25KK358 pKa = 7.83TSKK361 pKa = 9.95KK362 pKa = 9.95KK363 pKa = 7.8YY364 pKa = 7.47HH365 pKa = 5.97KK366 pKa = 9.95RR367 pKa = 11.84YY368 pKa = 7.74KK369 pKa = 10.15KK370 pKa = 9.84RR371 pKa = 11.84YY372 pKa = 8.78KK373 pKa = 10.27VYY375 pKa = 10.7KK376 pKa = 9.62PYY378 pKa = 10.61KK379 pKa = 8.37KK380 pKa = 9.95KK381 pKa = 10.76KK382 pKa = 8.5KK383 pKa = 9.57FRR385 pKa = 11.84SGKK388 pKa = 8.01YY389 pKa = 8.61FKK391 pKa = 10.48PKK393 pKa = 9.37EE394 pKa = 4.11KK395 pKa = 10.27KK396 pKa = 8.74GSKK399 pKa = 9.49RR400 pKa = 11.84KK401 pKa = 9.66YY402 pKa = 9.14CPKK405 pKa = 10.31GKK407 pKa = 9.77KK408 pKa = 9.42DD409 pKa = 3.64CRR411 pKa = 11.84CWICNIEE418 pKa = 3.72GHH420 pKa = 5.91YY421 pKa = 11.26ANEE424 pKa = 4.29CPNRR428 pKa = 11.84QSSEE432 pKa = 3.98KK433 pKa = 10.95AHH435 pKa = 6.69ILQQAEE441 pKa = 4.19NLGLQPVEE449 pKa = 4.07EE450 pKa = 4.93PYY452 pKa = 11.05EE453 pKa = 4.26GVQEE457 pKa = 4.31VFILEE462 pKa = 4.23YY463 pKa = 10.61KK464 pKa = 10.49EE465 pKa = 4.56EE466 pKa = 4.1EE467 pKa = 4.44EE468 pKa = 4.21EE469 pKa = 4.39TSTEE473 pKa = 3.97EE474 pKa = 4.75SDD476 pKa = 5.02DD477 pKa = 3.84EE478 pKa = 4.97SSTSEE483 pKa = 5.36DD484 pKa = 3.35SDD486 pKa = 3.84SDD488 pKa = 3.58
MM1 pKa = 7.73AEE3 pKa = 4.5SILDD7 pKa = 3.48RR8 pKa = 11.84TINRR12 pKa = 11.84FWYY15 pKa = 10.5NLGEE19 pKa = 4.56DD20 pKa = 4.62CLSEE24 pKa = 4.25SQFDD28 pKa = 3.63LMIRR32 pKa = 11.84LMEE35 pKa = 4.36EE36 pKa = 4.14SLDD39 pKa = 3.54GDD41 pKa = 3.95QIIDD45 pKa = 3.92LTSLPSDD52 pKa = 3.49NLQVEE57 pKa = 4.7QVMTTTDD64 pKa = 3.35DD65 pKa = 4.2SISEE69 pKa = 4.02EE70 pKa = 4.57SEE72 pKa = 3.64FLLAIGEE79 pKa = 4.16ISEE82 pKa = 4.84DD83 pKa = 3.61EE84 pKa = 4.21SDD86 pKa = 3.66SGEE89 pKa = 4.04EE90 pKa = 4.03PEE92 pKa = 4.39FEE94 pKa = 4.26QVRR97 pKa = 11.84MDD99 pKa = 3.4RR100 pKa = 11.84TGGTEE105 pKa = 3.59IPKK108 pKa = 10.47EE109 pKa = 3.95EE110 pKa = 4.82DD111 pKa = 3.31GEE113 pKa = 4.73GPSRR117 pKa = 11.84YY118 pKa = 9.29NEE120 pKa = 4.24RR121 pKa = 11.84KK122 pKa = 9.81RR123 pKa = 11.84KK124 pKa = 7.47TPEE127 pKa = 3.29DD128 pKa = 4.14RR129 pKa = 11.84YY130 pKa = 10.74FPTQPKK136 pKa = 8.96TIPGQKK142 pKa = 6.7QTSMGMLNIDD152 pKa = 3.45CQINRR157 pKa = 11.84RR158 pKa = 11.84TLIDD162 pKa = 3.36DD163 pKa = 4.03WAAEE167 pKa = 3.64IGLIVKK173 pKa = 7.4TNRR176 pKa = 11.84EE177 pKa = 4.21DD178 pKa = 3.71YY179 pKa = 10.97LDD181 pKa = 4.59PEE183 pKa = 4.86TILLLMEE190 pKa = 5.35HH191 pKa = 6.19KK192 pKa = 9.65TSGIAKK198 pKa = 8.99EE199 pKa = 4.39LIRR202 pKa = 11.84NTRR205 pKa = 11.84WNRR208 pKa = 11.84TTGDD212 pKa = 3.23IIEE215 pKa = 4.15QVINAMYY222 pKa = 10.9TMFLGLNYY230 pKa = 10.01SDD232 pKa = 4.16NKK234 pKa = 9.76VAEE237 pKa = 4.95KK238 pKa = 9.96IDD240 pKa = 3.71EE241 pKa = 4.11QEE243 pKa = 3.87KK244 pKa = 10.81AKK246 pKa = 10.57IRR248 pKa = 11.84MTKK251 pKa = 10.09LQLFDD256 pKa = 2.96ICYY259 pKa = 10.29LEE261 pKa = 5.62EE262 pKa = 4.13FTCDD266 pKa = 3.39YY267 pKa = 9.76EE268 pKa = 5.13KK269 pKa = 11.56NMYY272 pKa = 7.76KK273 pKa = 9.52TEE275 pKa = 3.96MADD278 pKa = 3.35FPGYY282 pKa = 10.03INQYY286 pKa = 9.44LSKK289 pKa = 10.49IPIIGEE295 pKa = 3.82KK296 pKa = 10.07ALTRR300 pKa = 11.84FRR302 pKa = 11.84HH303 pKa = 5.1EE304 pKa = 4.41ANGTSIYY311 pKa = 10.98SLGFAAKK318 pKa = 9.37IVKK321 pKa = 10.16EE322 pKa = 4.25EE323 pKa = 3.98LSKK326 pKa = 10.93ICDD329 pKa = 3.44LSKK332 pKa = 10.64KK333 pKa = 8.32QKK335 pKa = 10.13KK336 pKa = 9.23LKK338 pKa = 10.43KK339 pKa = 9.92FNKK342 pKa = 8.29KK343 pKa = 9.46CCSIGEE349 pKa = 4.05ASVEE353 pKa = 3.97YY354 pKa = 10.54GGKK357 pKa = 8.25KK358 pKa = 7.83TSKK361 pKa = 9.95KK362 pKa = 9.95KK363 pKa = 7.8YY364 pKa = 7.47HH365 pKa = 5.97KK366 pKa = 9.95RR367 pKa = 11.84YY368 pKa = 7.74KK369 pKa = 10.15KK370 pKa = 9.84RR371 pKa = 11.84YY372 pKa = 8.78KK373 pKa = 10.27VYY375 pKa = 10.7KK376 pKa = 9.62PYY378 pKa = 10.61KK379 pKa = 8.37KK380 pKa = 9.95KK381 pKa = 10.76KK382 pKa = 8.5KK383 pKa = 9.57FRR385 pKa = 11.84SGKK388 pKa = 8.01YY389 pKa = 8.61FKK391 pKa = 10.48PKK393 pKa = 9.37EE394 pKa = 4.11KK395 pKa = 10.27KK396 pKa = 8.74GSKK399 pKa = 9.49RR400 pKa = 11.84KK401 pKa = 9.66YY402 pKa = 9.14CPKK405 pKa = 10.31GKK407 pKa = 9.77KK408 pKa = 9.42DD409 pKa = 3.64CRR411 pKa = 11.84CWICNIEE418 pKa = 3.72GHH420 pKa = 5.91YY421 pKa = 11.26ANEE424 pKa = 4.29CPNRR428 pKa = 11.84QSSEE432 pKa = 3.98KK433 pKa = 10.95AHH435 pKa = 6.69ILQQAEE441 pKa = 4.19NLGLQPVEE449 pKa = 4.07EE450 pKa = 4.93PYY452 pKa = 11.05EE453 pKa = 4.26GVQEE457 pKa = 4.31VFILEE462 pKa = 4.23YY463 pKa = 10.61KK464 pKa = 10.49EE465 pKa = 4.56EE466 pKa = 4.1EE467 pKa = 4.44EE468 pKa = 4.21EE469 pKa = 4.39TSTEE473 pKa = 3.97EE474 pKa = 4.75SDD476 pKa = 5.02DD477 pKa = 3.84EE478 pKa = 4.97SSTSEE483 pKa = 5.36DD484 pKa = 3.35SDD486 pKa = 3.84SDD488 pKa = 3.58
Molecular weight: 56.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03552|VAP_CAMVC Virion-associated protein OS=Cauliflower mosaic virus (strain CM-1841) OX=10644 GN=ORF III PE=3 SV=1
MM1 pKa = 7.71SITGQPHH8 pKa = 6.08VYY10 pKa = 10.34KK11 pKa = 10.15KK12 pKa = 10.26DD13 pKa = 3.28TIIRR17 pKa = 11.84LKK19 pKa = 10.51PLSLNSNNRR28 pKa = 11.84SYY30 pKa = 11.62VFSSSKK36 pKa = 11.2GNIQNIINHH45 pKa = 6.59LNNLNEE51 pKa = 3.99IVGRR55 pKa = 11.84SLLGIWKK62 pKa = 9.14INSYY66 pKa = 10.68FGLSKK71 pKa = 10.89DD72 pKa = 3.5PSEE75 pKa = 4.67SKK77 pKa = 10.77SKK79 pKa = 10.86NPSVFNTAKK88 pKa = 10.78NIFKK92 pKa = 10.78SRR94 pKa = 11.84GVDD97 pKa = 3.2YY98 pKa = 10.92SSQLKK103 pKa = 8.92EE104 pKa = 4.12VKK106 pKa = 10.33SLLEE110 pKa = 4.07AQNTRR115 pKa = 11.84IKK117 pKa = 10.7NLEE120 pKa = 3.9NAIQSLDD127 pKa = 3.51NKK129 pKa = 10.23IEE131 pKa = 4.11PEE133 pKa = 4.1PLTKK137 pKa = 10.77EE138 pKa = 3.86EE139 pKa = 4.05VKK141 pKa = 10.52EE142 pKa = 3.89LKK144 pKa = 10.55EE145 pKa = 4.32SINSIKK151 pKa = 10.61EE152 pKa = 3.7GLKK155 pKa = 10.77NIIGG159 pKa = 3.69
MM1 pKa = 7.71SITGQPHH8 pKa = 6.08VYY10 pKa = 10.34KK11 pKa = 10.15KK12 pKa = 10.26DD13 pKa = 3.28TIIRR17 pKa = 11.84LKK19 pKa = 10.51PLSLNSNNRR28 pKa = 11.84SYY30 pKa = 11.62VFSSSKK36 pKa = 11.2GNIQNIINHH45 pKa = 6.59LNNLNEE51 pKa = 3.99IVGRR55 pKa = 11.84SLLGIWKK62 pKa = 9.14INSYY66 pKa = 10.68FGLSKK71 pKa = 10.89DD72 pKa = 3.5PSEE75 pKa = 4.67SKK77 pKa = 10.77SKK79 pKa = 10.86NPSVFNTAKK88 pKa = 10.78NIFKK92 pKa = 10.78SRR94 pKa = 11.84GVDD97 pKa = 3.2YY98 pKa = 10.92SSQLKK103 pKa = 8.92EE104 pKa = 4.12VKK106 pKa = 10.33SLLEE110 pKa = 4.07AQNTRR115 pKa = 11.84IKK117 pKa = 10.7NLEE120 pKa = 3.9NAIQSLDD127 pKa = 3.51NKK129 pKa = 10.23IEE131 pKa = 4.11PEE133 pKa = 4.1PLTKK137 pKa = 10.77EE138 pKa = 3.86EE139 pKa = 4.05VKK141 pKa = 10.52EE142 pKa = 3.89LKK144 pKa = 10.55EE145 pKa = 4.32SINSIKK151 pKa = 10.61EE152 pKa = 3.7GLKK155 pKa = 10.77NIIGG159 pKa = 3.69
Molecular weight: 17.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2302 |
129 |
679 |
383.7 |
43.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.039 ± 0.733 | 1.564 ± 0.273 |
5.3 ± 0.511 | 8.036 ± 1.12 |
3.779 ± 0.583 | 5.083 ± 0.373 |
2.042 ± 0.488 | 8.08 ± 0.428 |
10.556 ± 0.457 | 8.558 ± 0.598 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.215 ± 0.244 | 5.995 ± 1.141 |
4.605 ± 0.662 | 4.605 ± 0.316 |
3.736 ± 0.454 | 7.124 ± 0.838 |
5.343 ± 0.559 | 4.387 ± 0.579 |
0.869 ± 0.168 | 3.084 ± 0.405 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
