
Kallithea virus
Taxonomy: Viruses; Naldaviricetes; Lefavirales; Nudiviridae; Alphanudivirus; Drosophila melanogaster nudivirus A
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
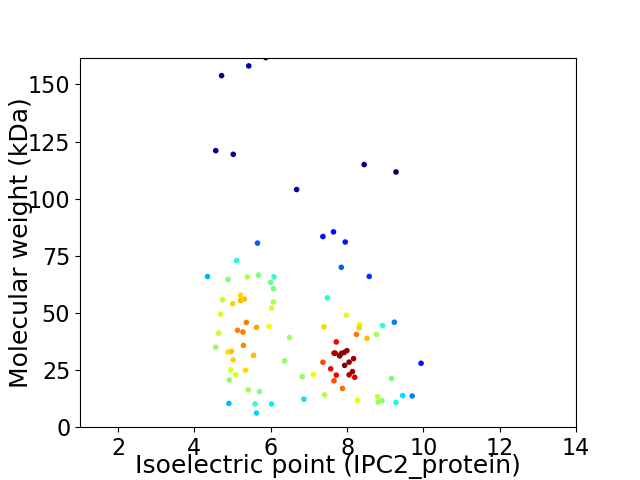
Virtual 2D-PAGE plot for 95 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
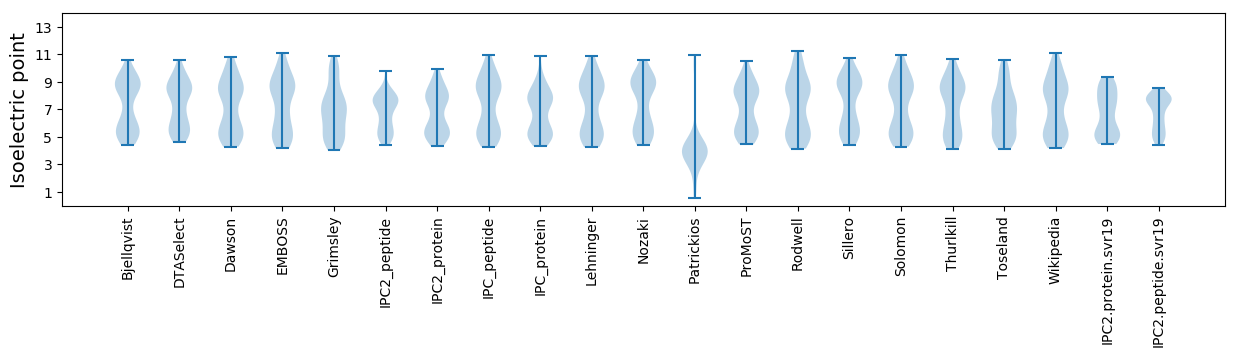
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S5VFX8|A0A1S5VFX8_9VIRU Uncharacterized protein OS=Kallithea virus OX=1654582 PE=4 SV=1
MM1 pKa = 7.23NVADD5 pKa = 4.55TPIGYY10 pKa = 8.91PNGLDD15 pKa = 3.43PNIQKK20 pKa = 9.79SQNVDD25 pKa = 3.49NIEE28 pKa = 4.27SGVTSINNNIDD39 pKa = 2.93NTTRR43 pKa = 11.84QRR45 pKa = 11.84ILYY48 pKa = 8.48ALQRR52 pKa = 11.84AIPDD56 pKa = 3.36LPKK59 pKa = 10.31IITEE63 pKa = 4.16LTNQKK68 pKa = 8.49HH69 pKa = 4.45TTPEE73 pKa = 3.94FEE75 pKa = 5.13LNNIVDD81 pKa = 3.99SLIGHH86 pKa = 5.5VQIICSQPASLYY98 pKa = 10.08TEE100 pKa = 3.9NDD102 pKa = 2.91YY103 pKa = 11.4KK104 pKa = 11.21LRR106 pKa = 11.84VLNAEE111 pKa = 3.34IDD113 pKa = 4.15YY114 pKa = 11.24VKK116 pKa = 10.57EE117 pKa = 4.12LYY119 pKa = 9.64IQNSGGLQSAIDD131 pKa = 4.04DD132 pKa = 4.27LKK134 pKa = 11.44VKK136 pKa = 10.56YY137 pKa = 10.27SNCSRR142 pKa = 11.84LLDD145 pKa = 3.84NATKK149 pKa = 10.52SLYY152 pKa = 8.7RR153 pKa = 11.84TNADD157 pKa = 3.61LNNLNVSQKK166 pKa = 10.78NSDD169 pKa = 3.65EE170 pKa = 4.14LTTQITTLQAEE181 pKa = 4.5AVNLKK186 pKa = 10.38NVISQMQTEE195 pKa = 4.57RR196 pKa = 11.84DD197 pKa = 3.47QMLKK201 pKa = 10.74SMQEE205 pKa = 3.43MSEE208 pKa = 4.1INEE211 pKa = 3.84SGQRR215 pKa = 11.84QKK217 pKa = 10.89ILRR220 pKa = 11.84SEE222 pKa = 3.88EE223 pKa = 3.77AQAKK227 pKa = 6.94EE228 pKa = 3.87RR229 pKa = 11.84RR230 pKa = 11.84DD231 pKa = 3.18IGILLRR237 pKa = 11.84VINPEE242 pKa = 4.11STSSDD247 pKa = 2.85IDD249 pKa = 3.46EE250 pKa = 4.9MLTQIHH256 pKa = 7.09EE257 pKa = 4.33YY258 pKa = 10.48LISTNTLRR266 pKa = 11.84NTIEE270 pKa = 4.0KK271 pKa = 10.45LEE273 pKa = 4.34RR274 pKa = 11.84DD275 pKa = 3.7LTVSSNKK282 pKa = 9.88YY283 pKa = 10.11KK284 pKa = 9.66EE285 pKa = 4.22TSDD288 pKa = 5.01RR289 pKa = 11.84LIDD292 pKa = 4.15CANALDD298 pKa = 4.0KK299 pKa = 11.84NEE301 pKa = 4.18TEE303 pKa = 4.11KK304 pKa = 11.31SKK306 pKa = 10.97LQFNFEE312 pKa = 4.28GKK314 pKa = 9.65IKK316 pKa = 10.5SLRR319 pKa = 11.84DD320 pKa = 3.32EE321 pKa = 4.26NAKK324 pKa = 10.87LMVTNSNAQAANDD337 pKa = 4.44DD338 pKa = 3.71LTSIYY343 pKa = 10.43EE344 pKa = 4.21RR345 pKa = 11.84TSSRR349 pKa = 11.84IKK351 pKa = 10.66DD352 pKa = 3.34IIDD355 pKa = 3.3NSKK358 pKa = 10.05PLSTDD363 pKa = 3.12DD364 pKa = 3.71VNSKK368 pKa = 10.15FLHH371 pKa = 6.15NLLQVLYY378 pKa = 10.24NEE380 pKa = 4.4NLSFVVPSDD389 pKa = 3.19ILIYY393 pKa = 9.57PQNPTANDD401 pKa = 3.16NDD403 pKa = 4.21AEE405 pKa = 4.41LVTIIDD411 pKa = 3.75SDD413 pKa = 4.12NNDD416 pKa = 3.16GDD418 pKa = 5.04GGIEE422 pKa = 5.01DD423 pKa = 5.22DD424 pKa = 4.16NNVDD428 pKa = 4.28NEE430 pKa = 4.18VEE432 pKa = 4.49YY433 pKa = 11.12YY434 pKa = 10.57SSQIPRR440 pKa = 11.84TPGSNIKK447 pKa = 10.68DD448 pKa = 3.69LDD450 pKa = 3.51QLISINSPEE459 pKa = 4.18TPPLVSASSSPVNAIDD475 pKa = 4.68DD476 pKa = 3.81SSIYY480 pKa = 10.44LISRR484 pKa = 11.84DD485 pKa = 4.11LPPTPTIPILDD496 pKa = 4.11SPTISTSNSPMDD508 pKa = 3.35EE509 pKa = 4.43SYY511 pKa = 10.48TRR513 pKa = 11.84RR514 pKa = 11.84PLKK517 pKa = 10.57RR518 pKa = 11.84RR519 pKa = 11.84TSDD522 pKa = 2.8INEE525 pKa = 4.2YY526 pKa = 10.7EE527 pKa = 4.18SVARR531 pKa = 11.84KK532 pKa = 9.63RR533 pKa = 11.84NQNEE537 pKa = 4.09TVSEE541 pKa = 4.36VTTEE545 pKa = 4.27VIDD548 pKa = 3.93EE549 pKa = 4.95LIITPQSNVVNRR561 pKa = 11.84DD562 pKa = 3.12DD563 pKa = 6.18DD564 pKa = 4.71DD565 pKa = 4.71FDD567 pKa = 6.35NIITINNDD575 pKa = 3.11DD576 pKa = 4.99DD577 pKa = 6.11SNLSNVNNVNN587 pKa = 3.16
MM1 pKa = 7.23NVADD5 pKa = 4.55TPIGYY10 pKa = 8.91PNGLDD15 pKa = 3.43PNIQKK20 pKa = 9.79SQNVDD25 pKa = 3.49NIEE28 pKa = 4.27SGVTSINNNIDD39 pKa = 2.93NTTRR43 pKa = 11.84QRR45 pKa = 11.84ILYY48 pKa = 8.48ALQRR52 pKa = 11.84AIPDD56 pKa = 3.36LPKK59 pKa = 10.31IITEE63 pKa = 4.16LTNQKK68 pKa = 8.49HH69 pKa = 4.45TTPEE73 pKa = 3.94FEE75 pKa = 5.13LNNIVDD81 pKa = 3.99SLIGHH86 pKa = 5.5VQIICSQPASLYY98 pKa = 10.08TEE100 pKa = 3.9NDD102 pKa = 2.91YY103 pKa = 11.4KK104 pKa = 11.21LRR106 pKa = 11.84VLNAEE111 pKa = 3.34IDD113 pKa = 4.15YY114 pKa = 11.24VKK116 pKa = 10.57EE117 pKa = 4.12LYY119 pKa = 9.64IQNSGGLQSAIDD131 pKa = 4.04DD132 pKa = 4.27LKK134 pKa = 11.44VKK136 pKa = 10.56YY137 pKa = 10.27SNCSRR142 pKa = 11.84LLDD145 pKa = 3.84NATKK149 pKa = 10.52SLYY152 pKa = 8.7RR153 pKa = 11.84TNADD157 pKa = 3.61LNNLNVSQKK166 pKa = 10.78NSDD169 pKa = 3.65EE170 pKa = 4.14LTTQITTLQAEE181 pKa = 4.5AVNLKK186 pKa = 10.38NVISQMQTEE195 pKa = 4.57RR196 pKa = 11.84DD197 pKa = 3.47QMLKK201 pKa = 10.74SMQEE205 pKa = 3.43MSEE208 pKa = 4.1INEE211 pKa = 3.84SGQRR215 pKa = 11.84QKK217 pKa = 10.89ILRR220 pKa = 11.84SEE222 pKa = 3.88EE223 pKa = 3.77AQAKK227 pKa = 6.94EE228 pKa = 3.87RR229 pKa = 11.84RR230 pKa = 11.84DD231 pKa = 3.18IGILLRR237 pKa = 11.84VINPEE242 pKa = 4.11STSSDD247 pKa = 2.85IDD249 pKa = 3.46EE250 pKa = 4.9MLTQIHH256 pKa = 7.09EE257 pKa = 4.33YY258 pKa = 10.48LISTNTLRR266 pKa = 11.84NTIEE270 pKa = 4.0KK271 pKa = 10.45LEE273 pKa = 4.34RR274 pKa = 11.84DD275 pKa = 3.7LTVSSNKK282 pKa = 9.88YY283 pKa = 10.11KK284 pKa = 9.66EE285 pKa = 4.22TSDD288 pKa = 5.01RR289 pKa = 11.84LIDD292 pKa = 4.15CANALDD298 pKa = 4.0KK299 pKa = 11.84NEE301 pKa = 4.18TEE303 pKa = 4.11KK304 pKa = 11.31SKK306 pKa = 10.97LQFNFEE312 pKa = 4.28GKK314 pKa = 9.65IKK316 pKa = 10.5SLRR319 pKa = 11.84DD320 pKa = 3.32EE321 pKa = 4.26NAKK324 pKa = 10.87LMVTNSNAQAANDD337 pKa = 4.44DD338 pKa = 3.71LTSIYY343 pKa = 10.43EE344 pKa = 4.21RR345 pKa = 11.84TSSRR349 pKa = 11.84IKK351 pKa = 10.66DD352 pKa = 3.34IIDD355 pKa = 3.3NSKK358 pKa = 10.05PLSTDD363 pKa = 3.12DD364 pKa = 3.71VNSKK368 pKa = 10.15FLHH371 pKa = 6.15NLLQVLYY378 pKa = 10.24NEE380 pKa = 4.4NLSFVVPSDD389 pKa = 3.19ILIYY393 pKa = 9.57PQNPTANDD401 pKa = 3.16NDD403 pKa = 4.21AEE405 pKa = 4.41LVTIIDD411 pKa = 3.75SDD413 pKa = 4.12NNDD416 pKa = 3.16GDD418 pKa = 5.04GGIEE422 pKa = 5.01DD423 pKa = 5.22DD424 pKa = 4.16NNVDD428 pKa = 4.28NEE430 pKa = 4.18VEE432 pKa = 4.49YY433 pKa = 11.12YY434 pKa = 10.57SSQIPRR440 pKa = 11.84TPGSNIKK447 pKa = 10.68DD448 pKa = 3.69LDD450 pKa = 3.51QLISINSPEE459 pKa = 4.18TPPLVSASSSPVNAIDD475 pKa = 4.68DD476 pKa = 3.81SSIYY480 pKa = 10.44LISRR484 pKa = 11.84DD485 pKa = 4.11LPPTPTIPILDD496 pKa = 4.11SPTISTSNSPMDD508 pKa = 3.35EE509 pKa = 4.43SYY511 pKa = 10.48TRR513 pKa = 11.84RR514 pKa = 11.84PLKK517 pKa = 10.57RR518 pKa = 11.84RR519 pKa = 11.84TSDD522 pKa = 2.8INEE525 pKa = 4.2YY526 pKa = 10.7EE527 pKa = 4.18SVARR531 pKa = 11.84KK532 pKa = 9.63RR533 pKa = 11.84NQNEE537 pKa = 4.09TVSEE541 pKa = 4.36VTTEE545 pKa = 4.27VIDD548 pKa = 3.93EE549 pKa = 4.95LIITPQSNVVNRR561 pKa = 11.84DD562 pKa = 3.12DD563 pKa = 6.18DD564 pKa = 4.71DD565 pKa = 4.71FDD567 pKa = 6.35NIITINNDD575 pKa = 3.11DD576 pKa = 4.99DD577 pKa = 6.11SNLSNVNNVNN587 pKa = 3.16
Molecular weight: 65.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7KLN5|A0A0F7KLN5_9VIRU ACH96169.1 GrBNV gp93-like protein OS=Kallithea virus OX=1654582 PE=4 SV=1
MM1 pKa = 7.2NVSIGNISEE10 pKa = 4.34FVNSIHH16 pKa = 8.01DD17 pKa = 4.43YY18 pKa = 10.43ILEE21 pKa = 4.23EE22 pKa = 4.3YY23 pKa = 10.24EE24 pKa = 3.93IIPRR28 pKa = 11.84DD29 pKa = 3.83FEE31 pKa = 4.4TKK33 pKa = 10.45LVIGGTAIAYY43 pKa = 7.83DD44 pKa = 4.08FNRR47 pKa = 11.84EE48 pKa = 3.66RR49 pKa = 11.84LIDD52 pKa = 4.09FDD54 pKa = 4.25CSKK57 pKa = 10.73LNKK60 pKa = 9.7HH61 pKa = 5.66IHH63 pKa = 4.44THH65 pKa = 6.11NDD67 pKa = 2.86VSHH70 pKa = 7.39DD71 pKa = 3.95KK72 pKa = 11.4YY73 pKa = 11.17LVCCSEE79 pKa = 4.31LDD81 pKa = 3.92IIALRR86 pKa = 11.84YY87 pKa = 10.01LNSCIFGPLKK97 pKa = 10.55VQIISTLHH105 pKa = 5.53HH106 pKa = 6.82WLPAATYY113 pKa = 7.83QHH115 pKa = 5.96VKK117 pKa = 10.63VSIQQRR123 pKa = 11.84GTLTINSYY131 pKa = 10.69FEE133 pKa = 4.03PSYY136 pKa = 11.09LFYY139 pKa = 11.14ALEE142 pKa = 4.01QSIGAHH148 pKa = 4.47IVYY151 pKa = 10.4NYY153 pKa = 8.55FHH155 pKa = 7.15SNIVTYY161 pKa = 10.83NSYY164 pKa = 10.99NLQGKK169 pKa = 7.87LTTIMVNARR178 pKa = 11.84RR179 pKa = 11.84RR180 pKa = 11.84SRR182 pKa = 11.84SRR184 pKa = 11.84SRR186 pKa = 11.84SASPARR192 pKa = 11.84RR193 pKa = 11.84SSRR196 pKa = 11.84ARR198 pKa = 11.84SPSPAGSKK206 pKa = 9.46RR207 pKa = 11.84RR208 pKa = 11.84RR209 pKa = 11.84PGRR212 pKa = 11.84PRR214 pKa = 11.84RR215 pKa = 11.84SSRR218 pKa = 11.84SGPRR222 pKa = 11.84PVAVARR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84SRR232 pKa = 11.84SRR234 pKa = 11.84SRR236 pKa = 11.84SRR238 pKa = 11.84SRR240 pKa = 11.84SRR242 pKa = 11.84AA243 pKa = 3.25
MM1 pKa = 7.2NVSIGNISEE10 pKa = 4.34FVNSIHH16 pKa = 8.01DD17 pKa = 4.43YY18 pKa = 10.43ILEE21 pKa = 4.23EE22 pKa = 4.3YY23 pKa = 10.24EE24 pKa = 3.93IIPRR28 pKa = 11.84DD29 pKa = 3.83FEE31 pKa = 4.4TKK33 pKa = 10.45LVIGGTAIAYY43 pKa = 7.83DD44 pKa = 4.08FNRR47 pKa = 11.84EE48 pKa = 3.66RR49 pKa = 11.84LIDD52 pKa = 4.09FDD54 pKa = 4.25CSKK57 pKa = 10.73LNKK60 pKa = 9.7HH61 pKa = 5.66IHH63 pKa = 4.44THH65 pKa = 6.11NDD67 pKa = 2.86VSHH70 pKa = 7.39DD71 pKa = 3.95KK72 pKa = 11.4YY73 pKa = 11.17LVCCSEE79 pKa = 4.31LDD81 pKa = 3.92IIALRR86 pKa = 11.84YY87 pKa = 10.01LNSCIFGPLKK97 pKa = 10.55VQIISTLHH105 pKa = 5.53HH106 pKa = 6.82WLPAATYY113 pKa = 7.83QHH115 pKa = 5.96VKK117 pKa = 10.63VSIQQRR123 pKa = 11.84GTLTINSYY131 pKa = 10.69FEE133 pKa = 4.03PSYY136 pKa = 11.09LFYY139 pKa = 11.14ALEE142 pKa = 4.01QSIGAHH148 pKa = 4.47IVYY151 pKa = 10.4NYY153 pKa = 8.55FHH155 pKa = 7.15SNIVTYY161 pKa = 10.83NSYY164 pKa = 10.99NLQGKK169 pKa = 7.87LTTIMVNARR178 pKa = 11.84RR179 pKa = 11.84RR180 pKa = 11.84SRR182 pKa = 11.84SRR184 pKa = 11.84SRR186 pKa = 11.84SASPARR192 pKa = 11.84RR193 pKa = 11.84SSRR196 pKa = 11.84ARR198 pKa = 11.84SPSPAGSKK206 pKa = 9.46RR207 pKa = 11.84RR208 pKa = 11.84RR209 pKa = 11.84PGRR212 pKa = 11.84PRR214 pKa = 11.84RR215 pKa = 11.84SSRR218 pKa = 11.84SGPRR222 pKa = 11.84PVAVARR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84SRR232 pKa = 11.84SRR234 pKa = 11.84SRR236 pKa = 11.84SRR238 pKa = 11.84SRR240 pKa = 11.84SRR242 pKa = 11.84AA243 pKa = 3.25
Molecular weight: 27.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
37704 |
53 |
1396 |
396.9 |
45.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.161 ± 0.159 | 1.841 ± 0.113 |
6.233 ± 0.254 | 4.835 ± 0.164 |
4.042 ± 0.159 | 3.451 ± 0.143 |
2.183 ± 0.109 | 8.718 ± 0.206 |
6.296 ± 0.231 | 8.031 ± 0.216 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.607 ± 0.104 | 8.649 ± 0.22 |
4.411 ± 0.203 | 4.339 ± 0.27 |
4.381 ± 0.144 | 8.272 ± 0.233 |
6.981 ± 0.326 | 5.339 ± 0.142 |
0.565 ± 0.044 | 4.665 ± 0.221 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
