
Muricauda sp.
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Muricauda; unclassified Muricauda
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
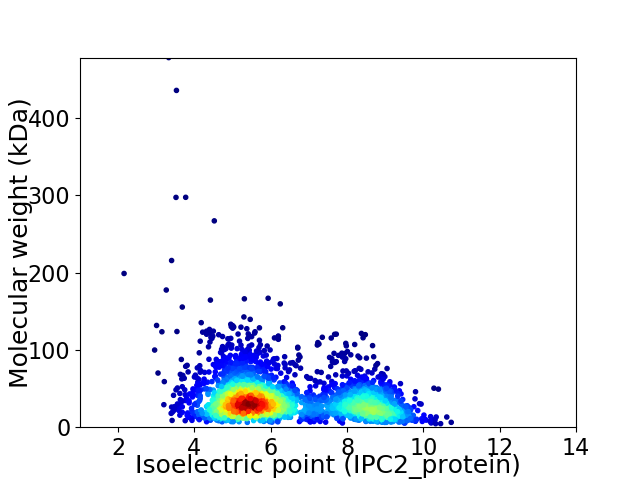
Virtual 2D-PAGE plot for 2810 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M8GJ28|A0A3M8GJ28_9FLAO Dihydroxy-acid dehydratase OS=Muricauda sp. OX=192149 GN=ilvD PE=3 SV=1
MM1 pKa = 7.65AGLFFVACSNDD12 pKa = 3.53DD13 pKa = 3.8DD14 pKa = 4.4ATGPTCTDD22 pKa = 4.75GIQNGDD28 pKa = 3.59EE29 pKa = 4.23TGVDD33 pKa = 3.85CGGSCAPCEE42 pKa = 4.08VAATCDD48 pKa = 4.06DD49 pKa = 5.12GIQNGDD55 pKa = 3.9EE56 pKa = 4.21EE57 pKa = 5.49GVDD60 pKa = 4.2CGGSSCSPCTTGIEE74 pKa = 4.05IPATYY79 pKa = 10.33VFEE82 pKa = 4.99RR83 pKa = 11.84NGEE86 pKa = 4.32STVSFSGQTTRR97 pKa = 11.84LLMARR102 pKa = 11.84EE103 pKa = 3.99VLGLLRR109 pKa = 11.84DD110 pKa = 3.99NTSTEE115 pKa = 3.67ADD117 pKa = 2.77IDD119 pKa = 3.79AAFDD123 pKa = 3.74HH124 pKa = 6.68QEE126 pKa = 3.79GAMDD130 pKa = 5.78FSDD133 pKa = 5.47DD134 pKa = 5.09DD135 pKa = 5.04LNASDD140 pKa = 3.54KK141 pKa = 10.26QVRR144 pKa = 11.84NKK146 pKa = 9.38TAGSYY151 pKa = 10.04VFSQLLPAVASIAVKK166 pKa = 10.68ADD168 pKa = 3.12FDD170 pKa = 4.42GYY172 pKa = 10.36IAEE175 pKa = 4.37QVNEE179 pKa = 4.69VFPFWNQVATEE190 pKa = 4.22GNAGQLPDD198 pKa = 3.94GSSTRR203 pKa = 11.84YY204 pKa = 9.06VNARR208 pKa = 11.84GLEE211 pKa = 3.88VDD213 pKa = 3.37QAFIKK218 pKa = 10.69GLTGAFVSDD227 pKa = 4.01QILNNYY233 pKa = 7.43TDD235 pKa = 4.5PGQLAQFEE243 pKa = 4.82DD244 pKa = 4.96DD245 pKa = 4.01NDD247 pKa = 3.62NDD249 pKa = 4.04VIVDD253 pKa = 4.23GKK255 pKa = 9.5TYY257 pKa = 10.88TDD259 pKa = 4.54MEE261 pKa = 4.9HH262 pKa = 7.29DD263 pKa = 3.17WDD265 pKa = 3.56EE266 pKa = 4.37AYY268 pKa = 10.52GYY270 pKa = 10.28FFGVAADD277 pKa = 4.04GEE279 pKa = 4.66NPVAALLDD287 pKa = 4.23GGDD290 pKa = 4.04PGDD293 pKa = 3.97FSSGSDD299 pKa = 3.18EE300 pKa = 4.48FLYY303 pKa = 10.56KK304 pKa = 10.79YY305 pKa = 10.55VDD307 pKa = 3.43NVEE310 pKa = 4.46ANFDD314 pKa = 3.62GTAAAIFDD322 pKa = 3.97AYY324 pKa = 10.02KK325 pKa = 10.52AGRR328 pKa = 11.84TAIVNKK334 pKa = 10.57DD335 pKa = 3.07YY336 pKa = 11.36DD337 pKa = 3.93EE338 pKa = 5.84RR339 pKa = 11.84DD340 pKa = 3.44AQADD344 pKa = 3.4ILRR347 pKa = 11.84EE348 pKa = 4.12KK349 pKa = 10.41IAMLVAVRR357 pKa = 11.84AIHH360 pKa = 5.46YY361 pKa = 8.28LKK363 pKa = 10.82AGASGVGGTNQGGTFHH379 pKa = 7.57DD380 pKa = 4.11LSEE383 pKa = 4.98AYY385 pKa = 10.21GFIYY389 pKa = 10.38SLQFLRR395 pKa = 11.84SPGSDD400 pKa = 2.63LAFYY404 pKa = 9.59FSRR407 pKa = 11.84DD408 pKa = 3.28EE409 pKa = 3.92VLGYY413 pKa = 10.77LDD415 pKa = 4.65TIYY418 pKa = 9.52NTPTNGFWNVTTEE431 pKa = 4.02NLDD434 pKa = 3.6EE435 pKa = 4.2VAEE438 pKa = 4.69AIAAKK443 pKa = 10.48FSFTAAEE450 pKa = 3.87VDD452 pKa = 3.42
MM1 pKa = 7.65AGLFFVACSNDD12 pKa = 3.53DD13 pKa = 3.8DD14 pKa = 4.4ATGPTCTDD22 pKa = 4.75GIQNGDD28 pKa = 3.59EE29 pKa = 4.23TGVDD33 pKa = 3.85CGGSCAPCEE42 pKa = 4.08VAATCDD48 pKa = 4.06DD49 pKa = 5.12GIQNGDD55 pKa = 3.9EE56 pKa = 4.21EE57 pKa = 5.49GVDD60 pKa = 4.2CGGSSCSPCTTGIEE74 pKa = 4.05IPATYY79 pKa = 10.33VFEE82 pKa = 4.99RR83 pKa = 11.84NGEE86 pKa = 4.32STVSFSGQTTRR97 pKa = 11.84LLMARR102 pKa = 11.84EE103 pKa = 3.99VLGLLRR109 pKa = 11.84DD110 pKa = 3.99NTSTEE115 pKa = 3.67ADD117 pKa = 2.77IDD119 pKa = 3.79AAFDD123 pKa = 3.74HH124 pKa = 6.68QEE126 pKa = 3.79GAMDD130 pKa = 5.78FSDD133 pKa = 5.47DD134 pKa = 5.09DD135 pKa = 5.04LNASDD140 pKa = 3.54KK141 pKa = 10.26QVRR144 pKa = 11.84NKK146 pKa = 9.38TAGSYY151 pKa = 10.04VFSQLLPAVASIAVKK166 pKa = 10.68ADD168 pKa = 3.12FDD170 pKa = 4.42GYY172 pKa = 10.36IAEE175 pKa = 4.37QVNEE179 pKa = 4.69VFPFWNQVATEE190 pKa = 4.22GNAGQLPDD198 pKa = 3.94GSSTRR203 pKa = 11.84YY204 pKa = 9.06VNARR208 pKa = 11.84GLEE211 pKa = 3.88VDD213 pKa = 3.37QAFIKK218 pKa = 10.69GLTGAFVSDD227 pKa = 4.01QILNNYY233 pKa = 7.43TDD235 pKa = 4.5PGQLAQFEE243 pKa = 4.82DD244 pKa = 4.96DD245 pKa = 4.01NDD247 pKa = 3.62NDD249 pKa = 4.04VIVDD253 pKa = 4.23GKK255 pKa = 9.5TYY257 pKa = 10.88TDD259 pKa = 4.54MEE261 pKa = 4.9HH262 pKa = 7.29DD263 pKa = 3.17WDD265 pKa = 3.56EE266 pKa = 4.37AYY268 pKa = 10.52GYY270 pKa = 10.28FFGVAADD277 pKa = 4.04GEE279 pKa = 4.66NPVAALLDD287 pKa = 4.23GGDD290 pKa = 4.04PGDD293 pKa = 3.97FSSGSDD299 pKa = 3.18EE300 pKa = 4.48FLYY303 pKa = 10.56KK304 pKa = 10.79YY305 pKa = 10.55VDD307 pKa = 3.43NVEE310 pKa = 4.46ANFDD314 pKa = 3.62GTAAAIFDD322 pKa = 3.97AYY324 pKa = 10.02KK325 pKa = 10.52AGRR328 pKa = 11.84TAIVNKK334 pKa = 10.57DD335 pKa = 3.07YY336 pKa = 11.36DD337 pKa = 3.93EE338 pKa = 5.84RR339 pKa = 11.84DD340 pKa = 3.44AQADD344 pKa = 3.4ILRR347 pKa = 11.84EE348 pKa = 4.12KK349 pKa = 10.41IAMLVAVRR357 pKa = 11.84AIHH360 pKa = 5.46YY361 pKa = 8.28LKK363 pKa = 10.82AGASGVGGTNQGGTFHH379 pKa = 7.57DD380 pKa = 4.11LSEE383 pKa = 4.98AYY385 pKa = 10.21GFIYY389 pKa = 10.38SLQFLRR395 pKa = 11.84SPGSDD400 pKa = 2.63LAFYY404 pKa = 9.59FSRR407 pKa = 11.84DD408 pKa = 3.28EE409 pKa = 3.92VLGYY413 pKa = 10.77LDD415 pKa = 4.65TIYY418 pKa = 9.52NTPTNGFWNVTTEE431 pKa = 4.02NLDD434 pKa = 3.6EE435 pKa = 4.2VAEE438 pKa = 4.69AIAAKK443 pKa = 10.48FSFTAAEE450 pKa = 3.87VDD452 pKa = 3.42
Molecular weight: 48.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M8GQ66|A0A3M8GQ66_9FLAO Uncharacterized protein OS=Muricauda sp. OX=192149 GN=ED555_04480 PE=4 SV=1
MM1 pKa = 7.63RR2 pKa = 11.84MFYY5 pKa = 10.79GILYY9 pKa = 7.91YY10 pKa = 9.77FQKK13 pKa = 10.55RR14 pKa = 11.84GFEE17 pKa = 3.99VCRR20 pKa = 11.84RR21 pKa = 11.84VAEE24 pKa = 4.14RR25 pKa = 11.84FGIRR29 pKa = 11.84ARR31 pKa = 11.84VVRR34 pKa = 11.84TTFIYY39 pKa = 9.47LTFATLGFGFALYY52 pKa = 10.87LFMAFWLRR60 pKa = 11.84IKK62 pKa = 10.7DD63 pKa = 4.08LIYY66 pKa = 10.52TKK68 pKa = 9.75RR69 pKa = 11.84TSVFDD74 pKa = 3.66LL75 pKa = 4.18
MM1 pKa = 7.63RR2 pKa = 11.84MFYY5 pKa = 10.79GILYY9 pKa = 7.91YY10 pKa = 9.77FQKK13 pKa = 10.55RR14 pKa = 11.84GFEE17 pKa = 3.99VCRR20 pKa = 11.84RR21 pKa = 11.84VAEE24 pKa = 4.14RR25 pKa = 11.84FGIRR29 pKa = 11.84ARR31 pKa = 11.84VVRR34 pKa = 11.84TTFIYY39 pKa = 9.47LTFATLGFGFALYY52 pKa = 10.87LFMAFWLRR60 pKa = 11.84IKK62 pKa = 10.7DD63 pKa = 4.08LIYY66 pKa = 10.52TKK68 pKa = 9.75RR69 pKa = 11.84TSVFDD74 pKa = 3.66LL75 pKa = 4.18
Molecular weight: 9.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
964233 |
38 |
4548 |
343.1 |
38.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.125 ± 0.044 | 0.72 ± 0.016 |
5.763 ± 0.045 | 6.626 ± 0.051 |
5.248 ± 0.039 | 7.026 ± 0.05 |
1.887 ± 0.027 | 7.019 ± 0.037 |
6.861 ± 0.076 | 9.638 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.279 ± 0.026 | 5.5 ± 0.042 |
3.613 ± 0.03 | 3.65 ± 0.024 |
3.894 ± 0.035 | 6.104 ± 0.036 |
5.576 ± 0.065 | 6.535 ± 0.033 |
1.144 ± 0.019 | 3.793 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
