
Pseudomonas phage MR14
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
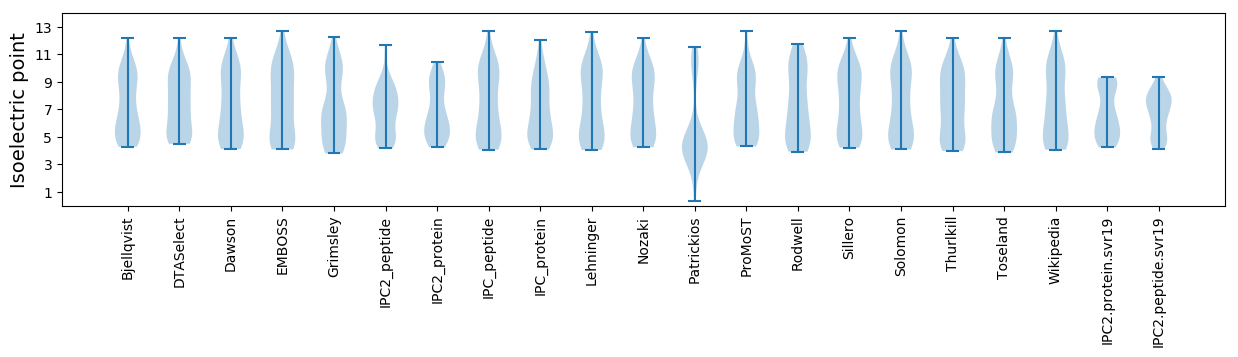
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
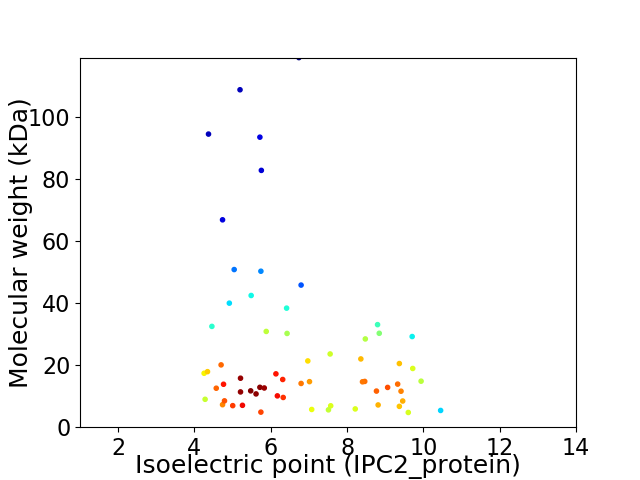
Virtual 2D-PAGE plot for 63 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
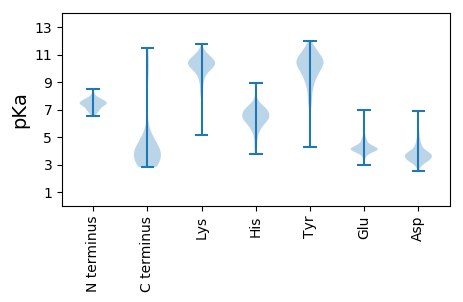
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M3TDR6|A0A6M3TDR6_9CAUD Uncharacterized protein OS=Pseudomonas phage MR14 OX=2711178 GN=Psm1vBMR14_gp27 PE=4 SV=1
MM1 pKa = 7.45ACEE4 pKa = 3.99TSAFLGRR11 pKa = 11.84DD12 pKa = 3.18TSLEE16 pKa = 3.93FAIGCGDD23 pKa = 3.75QLPAEE28 pKa = 4.84ADD30 pKa = 3.34WKK32 pKa = 11.07FIGALRR38 pKa = 11.84SNSLSGEE45 pKa = 4.07YY46 pKa = 9.45DD47 pKa = 3.66TVDD50 pKa = 3.2TTADD54 pKa = 3.58DD55 pKa = 3.64SAGGIRR61 pKa = 11.84EE62 pKa = 3.82MLATFKK68 pKa = 10.88SITISTDD75 pKa = 2.68GVARR79 pKa = 11.84RR80 pKa = 11.84ADD82 pKa = 3.5TATVNQTALLLHH94 pKa = 6.57YY95 pKa = 8.63YY96 pKa = 10.55NPVATGGQPVVWLRR110 pKa = 11.84RR111 pKa = 11.84TDD113 pKa = 3.44PRR115 pKa = 11.84ITLVGYY121 pKa = 9.96FLMSTYY127 pKa = 10.85DD128 pKa = 3.56LEE130 pKa = 4.46SPYY133 pKa = 11.36DD134 pKa = 3.75EE135 pKa = 4.84VSTFSIEE142 pKa = 4.28ASTTANSVIATGVSITATPVVV163 pKa = 3.62
MM1 pKa = 7.45ACEE4 pKa = 3.99TSAFLGRR11 pKa = 11.84DD12 pKa = 3.18TSLEE16 pKa = 3.93FAIGCGDD23 pKa = 3.75QLPAEE28 pKa = 4.84ADD30 pKa = 3.34WKK32 pKa = 11.07FIGALRR38 pKa = 11.84SNSLSGEE45 pKa = 4.07YY46 pKa = 9.45DD47 pKa = 3.66TVDD50 pKa = 3.2TTADD54 pKa = 3.58DD55 pKa = 3.64SAGGIRR61 pKa = 11.84EE62 pKa = 3.82MLATFKK68 pKa = 10.88SITISTDD75 pKa = 2.68GVARR79 pKa = 11.84RR80 pKa = 11.84ADD82 pKa = 3.5TATVNQTALLLHH94 pKa = 6.57YY95 pKa = 8.63YY96 pKa = 10.55NPVATGGQPVVWLRR110 pKa = 11.84RR111 pKa = 11.84TDD113 pKa = 3.44PRR115 pKa = 11.84ITLVGYY121 pKa = 9.96FLMSTYY127 pKa = 10.85DD128 pKa = 3.56LEE130 pKa = 4.46SPYY133 pKa = 11.36DD134 pKa = 3.75EE135 pKa = 4.84VSTFSIEE142 pKa = 4.28ASTTANSVIATGVSITATPVVV163 pKa = 3.62
Molecular weight: 17.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M3TDY3|A0A6M3TDY3_9CAUD Putative transcriptional regulator OS=Pseudomonas phage MR14 OX=2711178 GN=Psm1vBMR14_gp42c PE=4 SV=1
MM1 pKa = 7.53TGMPFWDD8 pKa = 4.02AYY10 pKa = 10.07KK11 pKa = 10.14PHH13 pKa = 7.46LYY15 pKa = 9.72NANRR19 pKa = 11.84RR20 pKa = 11.84VRR22 pKa = 11.84QRR24 pKa = 11.84QQRR27 pKa = 11.84TEE29 pKa = 3.9NPVSQRR35 pKa = 11.84YY36 pKa = 9.03AIRR39 pKa = 11.84ARR41 pKa = 11.84WGMEE45 pKa = 3.4WYY47 pKa = 9.73EE48 pKa = 4.14LLALLHH54 pKa = 6.11AQGLNRR60 pKa = 11.84QQCAQAMGYY69 pKa = 8.91SCWGSVKK76 pKa = 10.78AMLTRR81 pKa = 11.84HH82 pKa = 6.53PGNDD86 pKa = 3.1PFTPRR91 pKa = 11.84VLKK94 pKa = 10.11PKK96 pKa = 9.98RR97 pKa = 11.84QAPRR101 pKa = 11.84PRR103 pKa = 11.84FPRR106 pKa = 11.84RR107 pKa = 11.84HH108 pKa = 5.48AVEE111 pKa = 3.94QKK113 pKa = 9.82FGKK116 pKa = 10.21DD117 pKa = 3.13FWSVIRR123 pKa = 11.84DD124 pKa = 3.77FAEE127 pKa = 3.93KK128 pKa = 10.39DD129 pKa = 3.25YY130 pKa = 11.41TRR132 pKa = 11.84FDD134 pKa = 3.24TARR137 pKa = 11.84AIGCEE142 pKa = 3.67EE143 pKa = 4.12STFLRR148 pKa = 11.84ILADD152 pKa = 3.8NPKK155 pKa = 9.57HH156 pKa = 7.28DD157 pKa = 4.7PFEE160 pKa = 4.17SHH162 pKa = 6.38NVVGNYY168 pKa = 7.34TRR170 pKa = 11.84LTGEE174 pKa = 3.97SFEE177 pKa = 4.18AALRR181 pKa = 11.84RR182 pKa = 11.84MAAAGYY188 pKa = 8.86SRR190 pKa = 11.84RR191 pKa = 11.84QASIEE196 pKa = 3.54IGYY199 pKa = 9.22YY200 pKa = 9.93PSGLSLQRR208 pKa = 11.84AMDD211 pKa = 3.21IRR213 pKa = 11.84GIDD216 pKa = 3.46VTFKK220 pKa = 10.82KK221 pKa = 10.59AEE223 pKa = 4.08KK224 pKa = 9.33PKK226 pKa = 10.47KK227 pKa = 8.33PKK229 pKa = 10.12KK230 pKa = 9.44PRR232 pKa = 11.84EE233 pKa = 3.75PSISGRR239 pKa = 11.84YY240 pKa = 7.83GPKK243 pKa = 9.98SAIHH247 pKa = 6.61PWRR250 pKa = 11.84ISS252 pKa = 3.06
MM1 pKa = 7.53TGMPFWDD8 pKa = 4.02AYY10 pKa = 10.07KK11 pKa = 10.14PHH13 pKa = 7.46LYY15 pKa = 9.72NANRR19 pKa = 11.84RR20 pKa = 11.84VRR22 pKa = 11.84QRR24 pKa = 11.84QQRR27 pKa = 11.84TEE29 pKa = 3.9NPVSQRR35 pKa = 11.84YY36 pKa = 9.03AIRR39 pKa = 11.84ARR41 pKa = 11.84WGMEE45 pKa = 3.4WYY47 pKa = 9.73EE48 pKa = 4.14LLALLHH54 pKa = 6.11AQGLNRR60 pKa = 11.84QQCAQAMGYY69 pKa = 8.91SCWGSVKK76 pKa = 10.78AMLTRR81 pKa = 11.84HH82 pKa = 6.53PGNDD86 pKa = 3.1PFTPRR91 pKa = 11.84VLKK94 pKa = 10.11PKK96 pKa = 9.98RR97 pKa = 11.84QAPRR101 pKa = 11.84PRR103 pKa = 11.84FPRR106 pKa = 11.84RR107 pKa = 11.84HH108 pKa = 5.48AVEE111 pKa = 3.94QKK113 pKa = 9.82FGKK116 pKa = 10.21DD117 pKa = 3.13FWSVIRR123 pKa = 11.84DD124 pKa = 3.77FAEE127 pKa = 3.93KK128 pKa = 10.39DD129 pKa = 3.25YY130 pKa = 11.41TRR132 pKa = 11.84FDD134 pKa = 3.24TARR137 pKa = 11.84AIGCEE142 pKa = 3.67EE143 pKa = 4.12STFLRR148 pKa = 11.84ILADD152 pKa = 3.8NPKK155 pKa = 9.57HH156 pKa = 7.28DD157 pKa = 4.7PFEE160 pKa = 4.17SHH162 pKa = 6.38NVVGNYY168 pKa = 7.34TRR170 pKa = 11.84LTGEE174 pKa = 3.97SFEE177 pKa = 4.18AALRR181 pKa = 11.84RR182 pKa = 11.84MAAAGYY188 pKa = 8.86SRR190 pKa = 11.84RR191 pKa = 11.84QASIEE196 pKa = 3.54IGYY199 pKa = 9.22YY200 pKa = 9.93PSGLSLQRR208 pKa = 11.84AMDD211 pKa = 3.21IRR213 pKa = 11.84GIDD216 pKa = 3.46VTFKK220 pKa = 10.82KK221 pKa = 10.59AEE223 pKa = 4.08KK224 pKa = 9.33PKK226 pKa = 10.47KK227 pKa = 8.33PKK229 pKa = 10.12KK230 pKa = 9.44PRR232 pKa = 11.84EE233 pKa = 3.75PSISGRR239 pKa = 11.84YY240 pKa = 7.83GPKK243 pKa = 9.98SAIHH247 pKa = 6.61PWRR250 pKa = 11.84ISS252 pKa = 3.06
Molecular weight: 29.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14591 |
40 |
1079 |
231.6 |
25.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.857 ± 0.486 | 1.288 ± 0.178 |
5.49 ± 0.227 | 5.407 ± 0.323 |
3.098 ± 0.16 | 8.128 ± 0.259 |
1.816 ± 0.237 | 4.215 ± 0.17 |
4.14 ± 0.343 | 8.45 ± 0.204 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.316 ± 0.151 | 3.591 ± 0.226 |
5.14 ± 0.393 | 3.941 ± 0.264 |
6.189 ± 0.282 | 6.408 ± 0.284 |
6.895 ± 0.282 | 6.881 ± 0.234 |
1.864 ± 0.167 | 2.885 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
