
Sulfobacillus acidophilus (strain ATCC 700253 / DSM 10332 / NAL)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriales incertae sedis; Clostridiales Family XVII. Incertae Sedis; Sulfobacillus; Sulfobacillus acidophilus
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
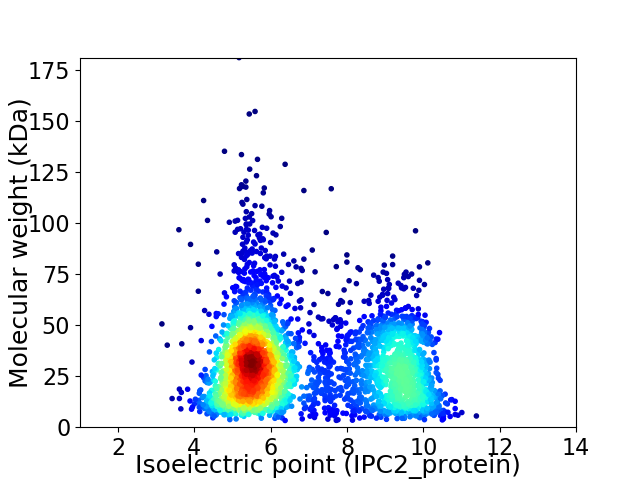
Virtual 2D-PAGE plot for 3444 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
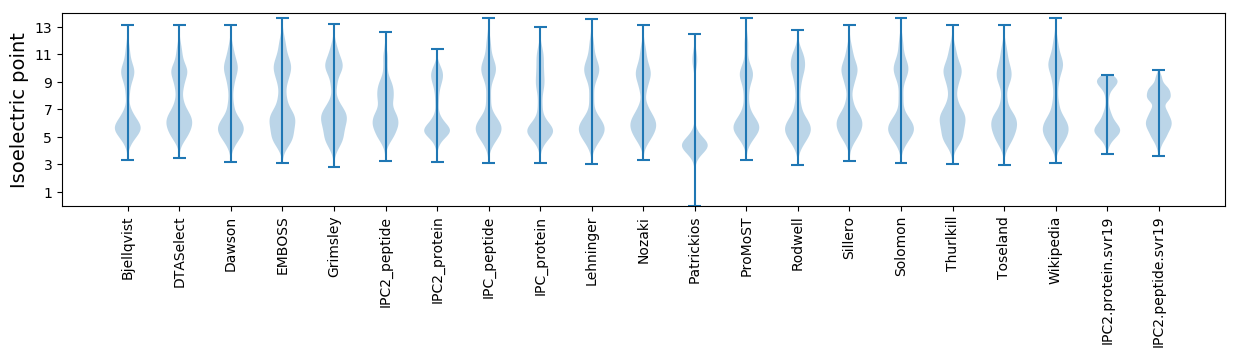
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8U176|G8U176_SULAD Uncharacterized protein OS=Sulfobacillus acidophilus (strain ATCC 700253 / DSM 10332 / NAL) OX=679936 GN=Sulac_0806 PE=4 SV=1
MM1 pKa = 7.6AFDD4 pKa = 4.16ILNIASRR11 pKa = 11.84AIAAQQLALEE21 pKa = 4.37VTGNNMANATTPGYY35 pKa = 9.72HH36 pKa = 6.06VEE38 pKa = 4.22SAVLAEE44 pKa = 4.88AMPSPDD50 pKa = 3.27PTEE53 pKa = 4.2PNALVGQGVDD63 pKa = 3.13VLTVSRR69 pKa = 11.84AANAFLSRR77 pKa = 11.84SVRR80 pKa = 11.84TQQSILGYY88 pKa = 10.21DD89 pKa = 3.33QAKK92 pKa = 9.68SQGLGEE98 pKa = 4.2VQNIFQEE105 pKa = 4.31PAPQGLAEE113 pKa = 4.16TMNAFFQSWLTLSEE127 pKa = 4.29NPTSVAAEE135 pKa = 3.98EE136 pKa = 5.14GVIQQGQNLSTVFNQMANTLTSQIASVNQNIGNLVTQVNQLATQIAQINQQIVTVGSSQEE196 pKa = 3.97PPNNLMDD203 pKa = 3.7QRR205 pKa = 11.84SALVDD210 pKa = 3.48QLSKK214 pKa = 10.96LVNISYY220 pKa = 10.99APGPDD225 pKa = 3.12NTMDD229 pKa = 3.62IYY231 pKa = 10.63IGTHH235 pKa = 5.92PLVSGNQASTLVTTANALGFNQPSWADD262 pKa = 3.22SGAPAAITSGTLGGNMALLYY282 pKa = 9.65TSSGSSGSPPGNGYY296 pKa = 8.22LTGYY300 pKa = 9.31LQSLNNLASAIVTQVNAQFNAGFTTSGTAPSQPFFVTTGTTAATIAVTPNLPPSDD355 pKa = 3.31IAAASSPNSPGDD367 pKa = 3.43GSNASAIFNLSQTAEE382 pKa = 4.63PIGSLTTTYY391 pKa = 9.85GQYY394 pKa = 8.94YY395 pKa = 5.92TTLVGQVGMDD405 pKa = 3.46GQSAEE410 pKa = 4.02NAANQDD416 pKa = 3.5QTTLTSLSNALQSATGVDD434 pKa = 3.55INQQSVQMIQEE445 pKa = 4.12QQSYY449 pKa = 7.7MAAAKK454 pKa = 9.92LVQTQQSIIQSLLAAVSS471 pKa = 3.45
MM1 pKa = 7.6AFDD4 pKa = 4.16ILNIASRR11 pKa = 11.84AIAAQQLALEE21 pKa = 4.37VTGNNMANATTPGYY35 pKa = 9.72HH36 pKa = 6.06VEE38 pKa = 4.22SAVLAEE44 pKa = 4.88AMPSPDD50 pKa = 3.27PTEE53 pKa = 4.2PNALVGQGVDD63 pKa = 3.13VLTVSRR69 pKa = 11.84AANAFLSRR77 pKa = 11.84SVRR80 pKa = 11.84TQQSILGYY88 pKa = 10.21DD89 pKa = 3.33QAKK92 pKa = 9.68SQGLGEE98 pKa = 4.2VQNIFQEE105 pKa = 4.31PAPQGLAEE113 pKa = 4.16TMNAFFQSWLTLSEE127 pKa = 4.29NPTSVAAEE135 pKa = 3.98EE136 pKa = 5.14GVIQQGQNLSTVFNQMANTLTSQIASVNQNIGNLVTQVNQLATQIAQINQQIVTVGSSQEE196 pKa = 3.97PPNNLMDD203 pKa = 3.7QRR205 pKa = 11.84SALVDD210 pKa = 3.48QLSKK214 pKa = 10.96LVNISYY220 pKa = 10.99APGPDD225 pKa = 3.12NTMDD229 pKa = 3.62IYY231 pKa = 10.63IGTHH235 pKa = 5.92PLVSGNQASTLVTTANALGFNQPSWADD262 pKa = 3.22SGAPAAITSGTLGGNMALLYY282 pKa = 9.65TSSGSSGSPPGNGYY296 pKa = 8.22LTGYY300 pKa = 9.31LQSLNNLASAIVTQVNAQFNAGFTTSGTAPSQPFFVTTGTTAATIAVTPNLPPSDD355 pKa = 3.31IAAASSPNSPGDD367 pKa = 3.43GSNASAIFNLSQTAEE382 pKa = 4.63PIGSLTTTYY391 pKa = 9.85GQYY394 pKa = 8.94YY395 pKa = 5.92TTLVGQVGMDD405 pKa = 3.46GQSAEE410 pKa = 4.02NAANQDD416 pKa = 3.5QTTLTSLSNALQSATGVDD434 pKa = 3.55INQQSVQMIQEE445 pKa = 4.12QQSYY449 pKa = 7.7MAAAKK454 pKa = 9.92LVQTQQSIIQSLLAAVSS471 pKa = 3.45
Molecular weight: 48.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8TV16|G8TV16_SULAD Uncharacterized protein OS=Sulfobacillus acidophilus (strain ATCC 700253 / DSM 10332 / NAL) OX=679936 GN=Sulac_0020 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.56RR3 pKa = 11.84TFQPNRR9 pKa = 11.84RR10 pKa = 11.84HH11 pKa = 5.61RR12 pKa = 11.84AKK14 pKa = 10.59VHH16 pKa = 5.69GFRR19 pKa = 11.84QRR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84QGRR28 pKa = 11.84AVLRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.22GRR39 pKa = 11.84KK40 pKa = 8.7RR41 pKa = 11.84LAGG44 pKa = 3.61
MM1 pKa = 7.44KK2 pKa = 9.56RR3 pKa = 11.84TFQPNRR9 pKa = 11.84RR10 pKa = 11.84HH11 pKa = 5.61RR12 pKa = 11.84AKK14 pKa = 10.59VHH16 pKa = 5.69GFRR19 pKa = 11.84QRR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84QGRR28 pKa = 11.84AVLRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QKK37 pKa = 10.22GRR39 pKa = 11.84KK40 pKa = 8.7RR41 pKa = 11.84LAGG44 pKa = 3.61
Molecular weight: 5.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1017646 |
30 |
1580 |
295.5 |
32.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.302 ± 0.044 | 0.553 ± 0.012 |
4.891 ± 0.031 | 5.347 ± 0.044 |
3.372 ± 0.028 | 8.19 ± 0.038 |
2.629 ± 0.019 | 5.23 ± 0.029 |
2.408 ± 0.025 | 10.78 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.35 ± 0.019 | 2.364 ± 0.025 |
5.951 ± 0.033 | 4.13 ± 0.027 |
7.253 ± 0.043 | 5.106 ± 0.029 |
5.744 ± 0.03 | 8.346 ± 0.033 |
2.363 ± 0.026 | 2.693 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
