
Algibacter pectinivorans
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Algibacter
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
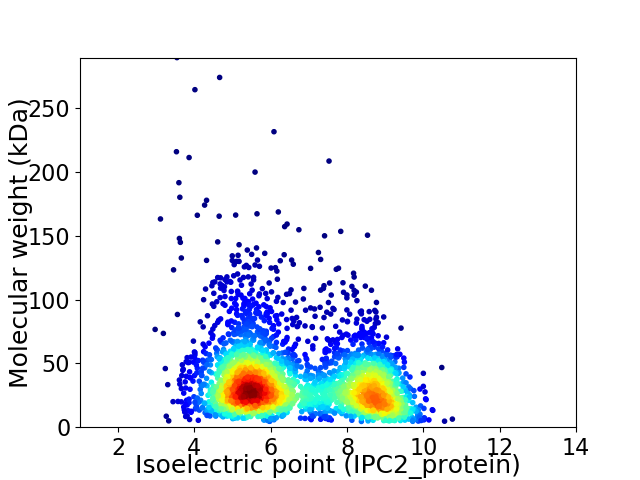
Virtual 2D-PAGE plot for 3126 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I1Q546|A0A1I1Q546_9FLAO TPM_phosphatase domain-containing protein OS=Algibacter pectinivorans OX=870482 GN=SAMN04487987_104232 PE=4 SV=1
MM1 pKa = 7.02KK2 pKa = 10.0TSLLFVVCIVMTIMSCNDD20 pKa = 3.06EE21 pKa = 5.3DD22 pKa = 4.74DD23 pKa = 4.05SQGINSTPEE32 pKa = 3.98FISFVDD38 pKa = 5.39AISSLPGEE46 pKa = 4.37TLTFEE51 pKa = 5.33AGISDD56 pKa = 3.82PAGIKK61 pKa = 10.11SINIKK66 pKa = 10.02YY67 pKa = 9.83EE68 pKa = 3.67SWFLDD73 pKa = 2.96KK74 pKa = 10.96TIFKK78 pKa = 10.66DD79 pKa = 3.52SLPDD83 pKa = 3.67VYY85 pKa = 11.0EE86 pKa = 3.77LAYY89 pKa = 10.79DD90 pKa = 4.6FIVPDD95 pKa = 4.2DD96 pKa = 4.32AEE98 pKa = 4.44MNSVHH103 pKa = 6.51TIVITAYY110 pKa = 10.51NVGGNQITKK119 pKa = 10.25NVVVTLDD126 pKa = 3.62KK127 pKa = 11.27DD128 pKa = 3.88VTNPTIQIVSPTNQSTVLIGDD149 pKa = 3.6GNEE152 pKa = 3.59IVLDD156 pKa = 3.62ITVSDD161 pKa = 3.97AEE163 pKa = 4.03LAEE166 pKa = 4.32FKK168 pKa = 10.59IEE170 pKa = 4.13SSVLNEE176 pKa = 3.77TKK178 pKa = 10.42AISGEE183 pKa = 4.11TYY185 pKa = 10.51NYY187 pKa = 9.63IRR189 pKa = 11.84ALDD192 pKa = 3.52ISTLGNYY199 pKa = 8.17EE200 pKa = 3.86FAITVTDD207 pKa = 3.5ASGNIRR213 pKa = 11.84TEE215 pKa = 4.28TISVNVLDD223 pKa = 4.24EE224 pKa = 4.41LLFEE228 pKa = 4.41VMYY231 pKa = 9.5ITDD234 pKa = 3.68VTSNAALNDD243 pKa = 3.82DD244 pKa = 4.57LFGVPYY250 pKa = 9.62STIASTASTEE260 pKa = 3.95DD261 pKa = 4.03GYY263 pKa = 11.96VFTGKK268 pKa = 10.48YY269 pKa = 9.9YY270 pKa = 10.83SSTPNSEE277 pKa = 3.65VRR279 pKa = 11.84FIPQQTAFEE288 pKa = 4.84PYY290 pKa = 10.6SFGADD295 pKa = 3.33PNAMGEE301 pKa = 4.38LILGNDD307 pKa = 3.61ASVSPIVVPGVGYY320 pKa = 9.9YY321 pKa = 10.11EE322 pKa = 3.81ITMDD326 pKa = 5.79LRR328 pKa = 11.84DD329 pKa = 3.23QSYY332 pKa = 8.99VVEE335 pKa = 4.74PYY337 pKa = 11.06VPIDD341 pKa = 3.27TAFDD345 pKa = 3.4QVYY348 pKa = 8.89VLGRR352 pKa = 11.84GVYY355 pKa = 10.39LDD357 pKa = 4.07ATSSTCVDD365 pKa = 3.34NSDD368 pKa = 2.95GSTRR372 pKa = 11.84CWHH375 pKa = 6.17FLSGKK380 pKa = 9.53PFIPDD385 pKa = 3.19SNNPYY390 pKa = 10.43LWTIDD395 pKa = 3.2VTAQDD400 pKa = 3.67QPDD403 pKa = 3.78DD404 pKa = 3.7NGANGFILNANPEE417 pKa = 3.86GWGPFWRR424 pKa = 11.84VDD426 pKa = 3.48ANDD429 pKa = 3.97PSVTIPNAGANYY441 pKa = 9.94VFDD444 pKa = 5.61DD445 pKa = 3.62SSLNKK450 pKa = 10.46DD451 pKa = 3.26YY452 pKa = 11.0TFVFDD457 pKa = 3.24THH459 pKa = 6.93LNRR462 pKa = 11.84LVVKK466 pKa = 10.66NRR468 pKa = 3.29
MM1 pKa = 7.02KK2 pKa = 10.0TSLLFVVCIVMTIMSCNDD20 pKa = 3.06EE21 pKa = 5.3DD22 pKa = 4.74DD23 pKa = 4.05SQGINSTPEE32 pKa = 3.98FISFVDD38 pKa = 5.39AISSLPGEE46 pKa = 4.37TLTFEE51 pKa = 5.33AGISDD56 pKa = 3.82PAGIKK61 pKa = 10.11SINIKK66 pKa = 10.02YY67 pKa = 9.83EE68 pKa = 3.67SWFLDD73 pKa = 2.96KK74 pKa = 10.96TIFKK78 pKa = 10.66DD79 pKa = 3.52SLPDD83 pKa = 3.67VYY85 pKa = 11.0EE86 pKa = 3.77LAYY89 pKa = 10.79DD90 pKa = 4.6FIVPDD95 pKa = 4.2DD96 pKa = 4.32AEE98 pKa = 4.44MNSVHH103 pKa = 6.51TIVITAYY110 pKa = 10.51NVGGNQITKK119 pKa = 10.25NVVVTLDD126 pKa = 3.62KK127 pKa = 11.27DD128 pKa = 3.88VTNPTIQIVSPTNQSTVLIGDD149 pKa = 3.6GNEE152 pKa = 3.59IVLDD156 pKa = 3.62ITVSDD161 pKa = 3.97AEE163 pKa = 4.03LAEE166 pKa = 4.32FKK168 pKa = 10.59IEE170 pKa = 4.13SSVLNEE176 pKa = 3.77TKK178 pKa = 10.42AISGEE183 pKa = 4.11TYY185 pKa = 10.51NYY187 pKa = 9.63IRR189 pKa = 11.84ALDD192 pKa = 3.52ISTLGNYY199 pKa = 8.17EE200 pKa = 3.86FAITVTDD207 pKa = 3.5ASGNIRR213 pKa = 11.84TEE215 pKa = 4.28TISVNVLDD223 pKa = 4.24EE224 pKa = 4.41LLFEE228 pKa = 4.41VMYY231 pKa = 9.5ITDD234 pKa = 3.68VTSNAALNDD243 pKa = 3.82DD244 pKa = 4.57LFGVPYY250 pKa = 9.62STIASTASTEE260 pKa = 3.95DD261 pKa = 4.03GYY263 pKa = 11.96VFTGKK268 pKa = 10.48YY269 pKa = 9.9YY270 pKa = 10.83SSTPNSEE277 pKa = 3.65VRR279 pKa = 11.84FIPQQTAFEE288 pKa = 4.84PYY290 pKa = 10.6SFGADD295 pKa = 3.33PNAMGEE301 pKa = 4.38LILGNDD307 pKa = 3.61ASVSPIVVPGVGYY320 pKa = 9.9YY321 pKa = 10.11EE322 pKa = 3.81ITMDD326 pKa = 5.79LRR328 pKa = 11.84DD329 pKa = 3.23QSYY332 pKa = 8.99VVEE335 pKa = 4.74PYY337 pKa = 11.06VPIDD341 pKa = 3.27TAFDD345 pKa = 3.4QVYY348 pKa = 8.89VLGRR352 pKa = 11.84GVYY355 pKa = 10.39LDD357 pKa = 4.07ATSSTCVDD365 pKa = 3.34NSDD368 pKa = 2.95GSTRR372 pKa = 11.84CWHH375 pKa = 6.17FLSGKK380 pKa = 9.53PFIPDD385 pKa = 3.19SNNPYY390 pKa = 10.43LWTIDD395 pKa = 3.2VTAQDD400 pKa = 3.67QPDD403 pKa = 3.78DD404 pKa = 3.7NGANGFILNANPEE417 pKa = 3.86GWGPFWRR424 pKa = 11.84VDD426 pKa = 3.48ANDD429 pKa = 3.97PSVTIPNAGANYY441 pKa = 9.94VFDD444 pKa = 5.61DD445 pKa = 3.62SSLNKK450 pKa = 10.46DD451 pKa = 3.26YY452 pKa = 11.0TFVFDD457 pKa = 3.24THH459 pKa = 6.93LNRR462 pKa = 11.84LVVKK466 pKa = 10.66NRR468 pKa = 3.29
Molecular weight: 51.31 kDa
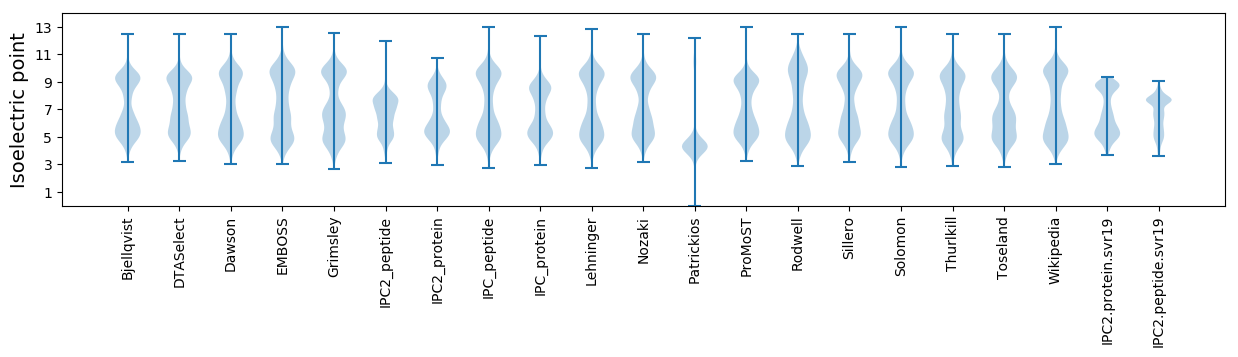
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I1QK21|A0A1I1QK21_9FLAO MJ0570-related uncharacterized domain-containing protein OS=Algibacter pectinivorans OX=870482 GN=SAMN04487987_106226 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
Molecular weight: 6.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1081701 |
39 |
2748 |
346.0 |
39.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.381 ± 0.039 | 0.771 ± 0.016 |
5.627 ± 0.039 | 6.312 ± 0.044 |
5.282 ± 0.038 | 6.261 ± 0.045 |
1.768 ± 0.021 | 7.971 ± 0.049 |
8.016 ± 0.059 | 9.246 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.031 ± 0.024 | 6.69 ± 0.051 |
3.313 ± 0.026 | 3.251 ± 0.024 |
3.203 ± 0.028 | 6.482 ± 0.034 |
5.996 ± 0.053 | 6.26 ± 0.032 |
1.042 ± 0.016 | 4.098 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
