
Banna-like virus strain Balaton/2010/HUN
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Seadornavirus; unclassified Seadornavirus
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
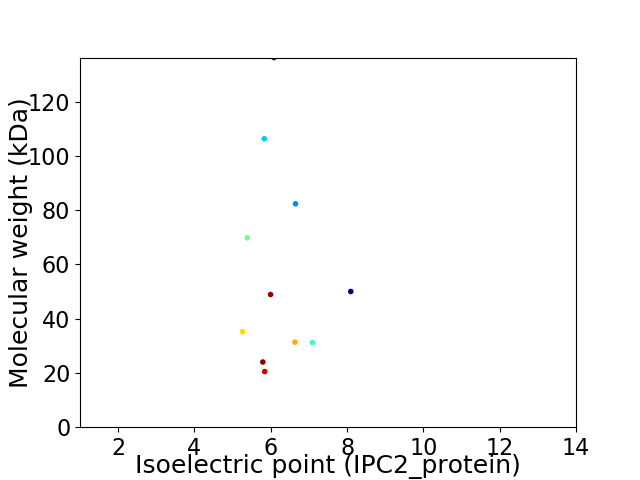
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|M9Z350|M9Z350_9REOV VP9 OS=Banna-like virus strain Balaton/2010/HUN OX=1325460 PE=4 SV=1
MM1 pKa = 7.58SGTKK5 pKa = 10.11RR6 pKa = 11.84IQLRR10 pKa = 11.84NGQGSIIKK18 pKa = 8.95TGQSYY23 pKa = 8.3EE24 pKa = 4.04LRR26 pKa = 11.84CRR28 pKa = 11.84RR29 pKa = 11.84LDD31 pKa = 3.46IDD33 pKa = 4.04CTLPLPGVTDD43 pKa = 3.26SRR45 pKa = 11.84GITDD49 pKa = 4.01SIKK52 pKa = 11.1YY53 pKa = 8.61IVEE56 pKa = 4.16YY57 pKa = 10.89LGLTYY62 pKa = 10.58VDD64 pKa = 4.15SNNGIGVDD72 pKa = 3.54EE73 pKa = 4.95RR74 pKa = 11.84DD75 pKa = 3.77FEE77 pKa = 4.68HH78 pKa = 6.8FQANPTLLVCRR89 pKa = 11.84TSIGPIFLKK98 pKa = 10.46KK99 pKa = 10.21LRR101 pKa = 11.84SLPVCKK107 pKa = 10.17CAFFEE112 pKa = 4.39SKK114 pKa = 11.19YY115 pKa = 10.93NILVGLKK122 pKa = 10.3GVIRR126 pKa = 11.84LRR128 pKa = 11.84NDD130 pKa = 2.95RR131 pKa = 11.84TGYY134 pKa = 9.82RR135 pKa = 11.84YY136 pKa = 10.06GVIMNQCFPPVINFTNFVEE155 pKa = 4.85AVCNLNYY162 pKa = 10.4LHH164 pKa = 6.95RR165 pKa = 11.84SEE167 pKa = 4.8VAVLHH172 pKa = 6.7GDD174 pKa = 3.73VNPGNIMSDD183 pKa = 3.25DD184 pKa = 3.95RR185 pKa = 11.84GCLKK189 pKa = 10.67LVDD192 pKa = 4.3PVCILEE198 pKa = 4.36GAVNMVNADD207 pKa = 3.74YY208 pKa = 11.45DD209 pKa = 4.31DD210 pKa = 4.19LTQACEE216 pKa = 3.87MKK218 pKa = 10.77VFIHH222 pKa = 6.33SLVILLAKK230 pKa = 9.77QFKK233 pKa = 9.85TSISEE238 pKa = 4.0LRR240 pKa = 11.84IDD242 pKa = 3.88YY243 pKa = 11.29SNVNPVFSTTKK254 pKa = 10.21GSPLEE259 pKa = 4.6SILDD263 pKa = 3.6YY264 pKa = 11.19SADD267 pKa = 5.41DD268 pKa = 4.1IISWKK273 pKa = 7.98RR274 pKa = 11.84TIMEE278 pKa = 4.38APALPTKK285 pKa = 10.9VNVFNDD291 pKa = 3.5EE292 pKa = 4.22YY293 pKa = 11.43YY294 pKa = 10.81RR295 pKa = 11.84LADD298 pKa = 3.72IPTMITEE305 pKa = 4.38NLDD308 pKa = 3.66DD309 pKa = 5.95DD310 pKa = 6.09DD311 pKa = 4.28YY312 pKa = 11.76TT313 pKa = 4.05
MM1 pKa = 7.58SGTKK5 pKa = 10.11RR6 pKa = 11.84IQLRR10 pKa = 11.84NGQGSIIKK18 pKa = 8.95TGQSYY23 pKa = 8.3EE24 pKa = 4.04LRR26 pKa = 11.84CRR28 pKa = 11.84RR29 pKa = 11.84LDD31 pKa = 3.46IDD33 pKa = 4.04CTLPLPGVTDD43 pKa = 3.26SRR45 pKa = 11.84GITDD49 pKa = 4.01SIKK52 pKa = 11.1YY53 pKa = 8.61IVEE56 pKa = 4.16YY57 pKa = 10.89LGLTYY62 pKa = 10.58VDD64 pKa = 4.15SNNGIGVDD72 pKa = 3.54EE73 pKa = 4.95RR74 pKa = 11.84DD75 pKa = 3.77FEE77 pKa = 4.68HH78 pKa = 6.8FQANPTLLVCRR89 pKa = 11.84TSIGPIFLKK98 pKa = 10.46KK99 pKa = 10.21LRR101 pKa = 11.84SLPVCKK107 pKa = 10.17CAFFEE112 pKa = 4.39SKK114 pKa = 11.19YY115 pKa = 10.93NILVGLKK122 pKa = 10.3GVIRR126 pKa = 11.84LRR128 pKa = 11.84NDD130 pKa = 2.95RR131 pKa = 11.84TGYY134 pKa = 9.82RR135 pKa = 11.84YY136 pKa = 10.06GVIMNQCFPPVINFTNFVEE155 pKa = 4.85AVCNLNYY162 pKa = 10.4LHH164 pKa = 6.95RR165 pKa = 11.84SEE167 pKa = 4.8VAVLHH172 pKa = 6.7GDD174 pKa = 3.73VNPGNIMSDD183 pKa = 3.25DD184 pKa = 3.95RR185 pKa = 11.84GCLKK189 pKa = 10.67LVDD192 pKa = 4.3PVCILEE198 pKa = 4.36GAVNMVNADD207 pKa = 3.74YY208 pKa = 11.45DD209 pKa = 4.31DD210 pKa = 4.19LTQACEE216 pKa = 3.87MKK218 pKa = 10.77VFIHH222 pKa = 6.33SLVILLAKK230 pKa = 9.77QFKK233 pKa = 9.85TSISEE238 pKa = 4.0LRR240 pKa = 11.84IDD242 pKa = 3.88YY243 pKa = 11.29SNVNPVFSTTKK254 pKa = 10.21GSPLEE259 pKa = 4.6SILDD263 pKa = 3.6YY264 pKa = 11.19SADD267 pKa = 5.41DD268 pKa = 4.1IISWKK273 pKa = 7.98RR274 pKa = 11.84TIMEE278 pKa = 4.38APALPTKK285 pKa = 10.9VNVFNDD291 pKa = 3.5EE292 pKa = 4.22YY293 pKa = 11.43YY294 pKa = 10.81RR295 pKa = 11.84LADD298 pKa = 3.72IPTMITEE305 pKa = 4.38NLDD308 pKa = 3.66DD309 pKa = 5.95DD310 pKa = 6.09DD311 pKa = 4.28YY312 pKa = 11.76TT313 pKa = 4.05
Molecular weight: 35.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M9Z5E3|M9Z5E3_9REOV VP6 OS=Banna-like virus strain Balaton/2010/HUN OX=1325460 PE=4 SV=1
MM1 pKa = 7.59EE2 pKa = 4.52IVRR5 pKa = 11.84TKK7 pKa = 10.4SQSAAITVGISQFFDD22 pKa = 3.22LTQNRR27 pKa = 11.84AVIEE31 pKa = 4.46TYY33 pKa = 10.75SEE35 pKa = 3.96FRR37 pKa = 11.84KK38 pKa = 10.33RR39 pKa = 11.84NAFILKK45 pKa = 9.73YY46 pKa = 10.48AEE48 pKa = 3.9QLFTGLPSLFKK59 pKa = 11.14DD60 pKa = 3.59EE61 pKa = 5.67LDD63 pKa = 3.52GLKK66 pKa = 10.31QIIEE70 pKa = 4.4SEE72 pKa = 4.03GGANSFAKK80 pKa = 8.82ITQVGLSHH88 pKa = 6.13MTQFLLSAKK97 pKa = 10.21LPADD101 pKa = 4.34DD102 pKa = 4.2YY103 pKa = 11.36LARR106 pKa = 11.84ALRR109 pKa = 11.84FIVFQIHH116 pKa = 6.03SNEE119 pKa = 3.75LRR121 pKa = 11.84KK122 pKa = 10.46GGALLMFDD130 pKa = 4.64NLLAYY135 pKa = 10.0LKK137 pKa = 10.55KK138 pKa = 10.04HH139 pKa = 5.4EE140 pKa = 4.31RR141 pKa = 11.84TNKK144 pKa = 9.42QSLISYY150 pKa = 6.79VTKK153 pKa = 10.75DD154 pKa = 3.66DD155 pKa = 3.31VDD157 pKa = 4.32IIIVNGKK164 pKa = 6.76TLTYY168 pKa = 10.11VCRR171 pKa = 11.84NADD174 pKa = 3.36ALKK177 pKa = 9.59TKK179 pKa = 10.01EE180 pKa = 3.76YY181 pKa = 11.06ANAEE185 pKa = 4.0GHH187 pKa = 6.38FVITDD192 pKa = 4.17TIGSFRR198 pKa = 11.84ASSSGMDD205 pKa = 2.95AVTFMTLKK213 pKa = 10.67SEE215 pKa = 4.23AEE217 pKa = 4.1HH218 pKa = 6.96IINLFGNAISIGQLYY233 pKa = 9.53YY234 pKa = 10.38RR235 pKa = 11.84QLHH238 pKa = 6.21AEE240 pKa = 4.07VSTLAKK246 pKa = 10.49LKK248 pKa = 10.32ANPGKK253 pKa = 8.19VHH255 pKa = 6.91RR256 pKa = 11.84MHH258 pKa = 6.94GNVRR262 pKa = 11.84DD263 pKa = 3.86GFVRR267 pKa = 11.84RR268 pKa = 11.84LNDD271 pKa = 3.43STKK274 pKa = 10.48HH275 pKa = 5.33RR276 pKa = 11.84VTGAQRR282 pKa = 11.84ALDD285 pKa = 3.61HH286 pKa = 6.94SNIIRR291 pKa = 11.84FKK293 pKa = 11.06SSGAGKK299 pKa = 9.71DD300 pKa = 3.23EE301 pKa = 4.26RR302 pKa = 11.84LTYY305 pKa = 10.75LVDD308 pKa = 3.4EE309 pKa = 5.21CFLKK313 pKa = 11.19SMLPDD318 pKa = 3.12NRR320 pKa = 11.84ACARR324 pKa = 11.84IDD326 pKa = 3.92NINALIASKK335 pKa = 10.5LLHH338 pKa = 6.66DD339 pKa = 4.71CKK341 pKa = 10.68IGTACMHH348 pKa = 6.66CGVIGSMDD356 pKa = 3.58TVSQLSEE363 pKa = 4.13RR364 pKa = 11.84GVDD367 pKa = 3.38IVANTNEE374 pKa = 4.49TLDD377 pKa = 3.78AMAAPVMTKK386 pKa = 10.31PEE388 pKa = 4.35AEE390 pKa = 4.38TVTDD394 pKa = 3.61ASLNARR400 pKa = 11.84IVQSKK405 pKa = 8.34GQSYY409 pKa = 9.54VPKK412 pKa = 10.06TEE414 pKa = 3.97YY415 pKa = 9.99KK416 pKa = 10.36AKK418 pKa = 10.24KK419 pKa = 9.93SEE421 pKa = 4.06PADD424 pKa = 3.12SGFTVKK430 pKa = 10.45VEE432 pKa = 4.51SNHH435 pKa = 5.93SVTDD439 pKa = 3.76ALVAGVVSRR448 pKa = 11.84EE449 pKa = 3.83KK450 pKa = 11.36LNFF453 pKa = 3.58
MM1 pKa = 7.59EE2 pKa = 4.52IVRR5 pKa = 11.84TKK7 pKa = 10.4SQSAAITVGISQFFDD22 pKa = 3.22LTQNRR27 pKa = 11.84AVIEE31 pKa = 4.46TYY33 pKa = 10.75SEE35 pKa = 3.96FRR37 pKa = 11.84KK38 pKa = 10.33RR39 pKa = 11.84NAFILKK45 pKa = 9.73YY46 pKa = 10.48AEE48 pKa = 3.9QLFTGLPSLFKK59 pKa = 11.14DD60 pKa = 3.59EE61 pKa = 5.67LDD63 pKa = 3.52GLKK66 pKa = 10.31QIIEE70 pKa = 4.4SEE72 pKa = 4.03GGANSFAKK80 pKa = 8.82ITQVGLSHH88 pKa = 6.13MTQFLLSAKK97 pKa = 10.21LPADD101 pKa = 4.34DD102 pKa = 4.2YY103 pKa = 11.36LARR106 pKa = 11.84ALRR109 pKa = 11.84FIVFQIHH116 pKa = 6.03SNEE119 pKa = 3.75LRR121 pKa = 11.84KK122 pKa = 10.46GGALLMFDD130 pKa = 4.64NLLAYY135 pKa = 10.0LKK137 pKa = 10.55KK138 pKa = 10.04HH139 pKa = 5.4EE140 pKa = 4.31RR141 pKa = 11.84TNKK144 pKa = 9.42QSLISYY150 pKa = 6.79VTKK153 pKa = 10.75DD154 pKa = 3.66DD155 pKa = 3.31VDD157 pKa = 4.32IIIVNGKK164 pKa = 6.76TLTYY168 pKa = 10.11VCRR171 pKa = 11.84NADD174 pKa = 3.36ALKK177 pKa = 9.59TKK179 pKa = 10.01EE180 pKa = 3.76YY181 pKa = 11.06ANAEE185 pKa = 4.0GHH187 pKa = 6.38FVITDD192 pKa = 4.17TIGSFRR198 pKa = 11.84ASSSGMDD205 pKa = 2.95AVTFMTLKK213 pKa = 10.67SEE215 pKa = 4.23AEE217 pKa = 4.1HH218 pKa = 6.96IINLFGNAISIGQLYY233 pKa = 9.53YY234 pKa = 10.38RR235 pKa = 11.84QLHH238 pKa = 6.21AEE240 pKa = 4.07VSTLAKK246 pKa = 10.49LKK248 pKa = 10.32ANPGKK253 pKa = 8.19VHH255 pKa = 6.91RR256 pKa = 11.84MHH258 pKa = 6.94GNVRR262 pKa = 11.84DD263 pKa = 3.86GFVRR267 pKa = 11.84RR268 pKa = 11.84LNDD271 pKa = 3.43STKK274 pKa = 10.48HH275 pKa = 5.33RR276 pKa = 11.84VTGAQRR282 pKa = 11.84ALDD285 pKa = 3.61HH286 pKa = 6.94SNIIRR291 pKa = 11.84FKK293 pKa = 11.06SSGAGKK299 pKa = 9.71DD300 pKa = 3.23EE301 pKa = 4.26RR302 pKa = 11.84LTYY305 pKa = 10.75LVDD308 pKa = 3.4EE309 pKa = 5.21CFLKK313 pKa = 11.19SMLPDD318 pKa = 3.12NRR320 pKa = 11.84ACARR324 pKa = 11.84IDD326 pKa = 3.92NINALIASKK335 pKa = 10.5LLHH338 pKa = 6.66DD339 pKa = 4.71CKK341 pKa = 10.68IGTACMHH348 pKa = 6.66CGVIGSMDD356 pKa = 3.58TVSQLSEE363 pKa = 4.13RR364 pKa = 11.84GVDD367 pKa = 3.38IVANTNEE374 pKa = 4.49TLDD377 pKa = 3.78AMAAPVMTKK386 pKa = 10.31PEE388 pKa = 4.35AEE390 pKa = 4.38TVTDD394 pKa = 3.61ASLNARR400 pKa = 11.84IVQSKK405 pKa = 8.34GQSYY409 pKa = 9.54VPKK412 pKa = 10.06TEE414 pKa = 3.97YY415 pKa = 9.99KK416 pKa = 10.36AKK418 pKa = 10.24KK419 pKa = 9.93SEE421 pKa = 4.06PADD424 pKa = 3.12SGFTVKK430 pKa = 10.45VEE432 pKa = 4.51SNHH435 pKa = 5.93SVTDD439 pKa = 3.76ALVAGVVSRR448 pKa = 11.84EE449 pKa = 3.83KK450 pKa = 11.36LNFF453 pKa = 3.58
Molecular weight: 50.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5704 |
182 |
1214 |
518.5 |
57.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.679 ± 0.623 | 1.192 ± 0.195 |
6.189 ± 0.289 | 4.786 ± 0.247 |
3.629 ± 0.222 | 5.575 ± 0.256 |
2.454 ± 0.286 | 6.697 ± 0.236 |
4.488 ± 0.357 | 9.309 ± 0.316 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.77 ± 0.152 | 5.54 ± 0.343 |
4.418 ± 0.32 | 3.384 ± 0.298 |
5.75 ± 0.329 | 7.626 ± 0.171 |
7.048 ± 0.408 | 6.96 ± 0.282 |
0.666 ± 0.143 | 3.839 ± 0.199 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
