
Pseudomonas caspiana
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; Pseudomonas syringae group
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
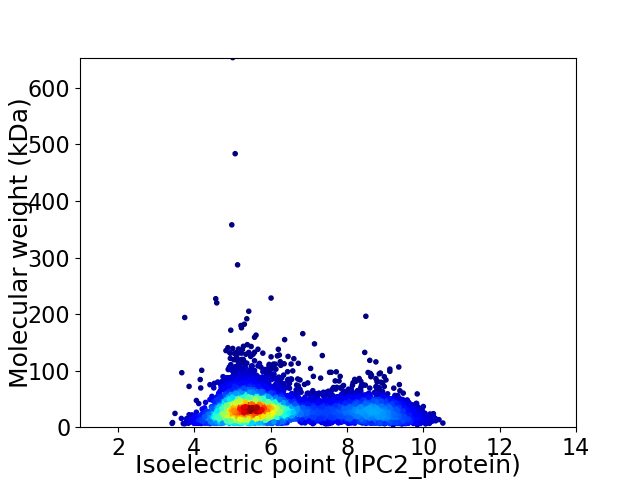
Virtual 2D-PAGE plot for 5280 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y3P468|A0A1Y3P468_9PSED Uncharacterized protein OS=Pseudomonas caspiana OX=1451454 GN=AUC60_07720 PE=4 SV=1
MM1 pKa = 7.43VGFIGTVRR9 pKa = 11.84QVVGEE14 pKa = 4.12VFAVAGDD21 pKa = 3.57GTRR24 pKa = 11.84RR25 pKa = 11.84LLIEE29 pKa = 4.32GDD31 pKa = 3.35RR32 pKa = 11.84LFAGEE37 pKa = 3.93QLQTGSGGAVAVNLDD52 pKa = 3.37AGGEE56 pKa = 4.15LTLGRR61 pKa = 11.84GSSLQLTPQLLNHH74 pKa = 6.01QAGHH78 pKa = 5.01VTTGDD83 pKa = 3.47EE84 pKa = 4.49LTPSLTPSSSTPAVLPEE101 pKa = 4.01QVFSADD107 pKa = 4.22APTQTPEE114 pKa = 3.6AAAPEE119 pKa = 4.24PAPDD123 pKa = 4.03GSSLGGGHH131 pKa = 6.44SAMLLTEE138 pKa = 4.12VGGEE142 pKa = 3.94VSPEE146 pKa = 3.54IGFTTQGLSSAPEE159 pKa = 3.87FPEE162 pKa = 4.16GRR164 pKa = 11.84IEE166 pKa = 4.2GLPDD170 pKa = 3.34SSDD173 pKa = 3.12ASGSVFVPPVVVVPPTEE190 pKa = 4.13PEE192 pKa = 4.06PPVEE196 pKa = 4.03PEE198 pKa = 4.15PPVEE202 pKa = 4.22HH203 pKa = 6.71PVDD206 pKa = 3.63HH207 pKa = 6.44GVEE210 pKa = 3.95LSGGQLTLAEE220 pKa = 5.17GDD222 pKa = 4.14LSCGPVTQSGVFTVNAVDD240 pKa = 4.25GLQSLVVGGLAVISGGVVAGFPLSTQTPLGNTFSVTAYY278 pKa = 10.47DD279 pKa = 3.72PATGEE284 pKa = 3.83VSYY287 pKa = 11.07SYY289 pKa = 9.63TLQGAIEE296 pKa = 4.29HH297 pKa = 6.52PVGEE301 pKa = 4.5GANTLGEE308 pKa = 4.42SFTVVARR315 pKa = 11.84DD316 pKa = 3.52TDD318 pKa = 3.73GDD320 pKa = 4.0QADD323 pKa = 3.76STLDD327 pKa = 3.25INIVDD332 pKa = 4.72DD333 pKa = 4.24VPEE336 pKa = 3.87AHH338 pKa = 7.04CIEE341 pKa = 4.47VTLSADD347 pKa = 3.81PDD349 pKa = 4.17CPGAPLPGASGSLLQGGTFGADD371 pKa = 2.48GGFIHH376 pKa = 7.13SISLNGTTYY385 pKa = 10.73SYY387 pKa = 11.4DD388 pKa = 3.59PEE390 pKa = 4.77TGCSTGSCEE399 pKa = 4.16GTGVFDD405 pKa = 4.04PATYY409 pKa = 10.71SLTVTSEE416 pKa = 3.97GGGTLVVNMLNGDD429 pKa = 4.24FSYY432 pKa = 10.84TPSTNPACEE441 pKa = 3.83PLTEE445 pKa = 4.6NIHH448 pKa = 6.05YY449 pKa = 9.96VISDD453 pKa = 3.56NDD455 pKa = 3.49GDD457 pKa = 4.35LAGSDD462 pKa = 3.87LQIHH466 pKa = 6.0VPAILPPEE474 pKa = 4.68GPTAVVDD481 pKa = 3.86NVITNILAPCIEE493 pKa = 4.46VPGVLLVTNDD503 pKa = 3.32VPGSGGPLTASPTVFHH519 pKa = 6.65TGWQVKK525 pKa = 10.63GEE527 pKa = 4.22GFSNDD532 pKa = 3.29CQKK535 pKa = 10.0TIDD538 pKa = 3.96FSGKK542 pKa = 9.3RR543 pKa = 11.84DD544 pKa = 3.42NSANQLKK551 pKa = 10.2NLEE554 pKa = 4.27RR555 pKa = 11.84SDD557 pKa = 3.98FANTGAMTALVVITGYY573 pKa = 10.9LGAFSGPGANAQDD586 pKa = 4.31LYY588 pKa = 11.35SVNLMAGEE596 pKa = 4.32SVAVDD601 pKa = 4.23LKK603 pKa = 11.49SLGDD607 pKa = 3.49QVGVFWQLDD616 pKa = 3.46GGEE619 pKa = 4.41FQPLDD624 pKa = 3.45AGGTFTATEE633 pKa = 3.89NGVYY637 pKa = 10.3RR638 pKa = 11.84LLVVNQPDD646 pKa = 3.45AGQPGPAVHH655 pKa = 5.73YY656 pKa = 9.45TLDD659 pKa = 3.7LVIDD663 pKa = 3.9YY664 pKa = 10.28SAADD668 pKa = 3.49TTPNHH673 pKa = 6.07EE674 pKa = 4.1GAYY677 pKa = 8.45TVNDD681 pKa = 3.52GHH683 pKa = 6.62GGSDD687 pKa = 3.09SAAVNISYY695 pKa = 10.58QEE697 pKa = 4.24GCTLLGTTGDD707 pKa = 3.95DD708 pKa = 3.55VLLGGAQDD716 pKa = 4.41DD717 pKa = 4.51SLHH720 pKa = 6.65GGDD723 pKa = 4.84GDD725 pKa = 4.46DD726 pKa = 4.09VLSGGAGNNDD736 pKa = 3.21LYY738 pKa = 11.37GDD740 pKa = 3.93NGNDD744 pKa = 3.37VLFSGSGNDD753 pKa = 3.67VLDD756 pKa = 4.39GGAGNNTASYY766 pKa = 10.87ALADD770 pKa = 3.6SGVTVSTAQLGPQHH784 pKa = 6.5TGGAGTDD791 pKa = 3.39TLINIQNLTGSDD803 pKa = 3.95YY804 pKa = 11.33DD805 pKa = 3.61DD806 pKa = 5.18HH807 pKa = 6.88LTGDD811 pKa = 3.74YY812 pKa = 8.44EE813 pKa = 5.1ANVINGGLGNDD824 pKa = 3.62VLVGGLGNDD833 pKa = 3.71TLTGGAGHH841 pKa = 7.22DD842 pKa = 3.69TFKK845 pKa = 10.78WLAGDD850 pKa = 3.84SGHH853 pKa = 7.49DD854 pKa = 3.6LVTDD858 pKa = 4.35FSLGADD864 pKa = 3.41TLDD867 pKa = 4.64LSKK870 pKa = 10.89LLQGFSVGTDD880 pKa = 3.14SLDD883 pKa = 3.66DD884 pKa = 3.86FLHH887 pKa = 6.36FKK889 pKa = 9.13VTGSGADD896 pKa = 3.33LVSSIEE902 pKa = 3.98IGSGGSSQTIDD913 pKa = 3.25LAGVNLAQHH922 pKa = 6.34YY923 pKa = 9.9DD924 pKa = 3.5VGVGAGGIVAGADD937 pKa = 3.02AGSIISGMLGDD948 pKa = 3.91HH949 pKa = 6.4SLRR952 pKa = 11.84ADD954 pKa = 3.56VVV956 pKa = 3.47
MM1 pKa = 7.43VGFIGTVRR9 pKa = 11.84QVVGEE14 pKa = 4.12VFAVAGDD21 pKa = 3.57GTRR24 pKa = 11.84RR25 pKa = 11.84LLIEE29 pKa = 4.32GDD31 pKa = 3.35RR32 pKa = 11.84LFAGEE37 pKa = 3.93QLQTGSGGAVAVNLDD52 pKa = 3.37AGGEE56 pKa = 4.15LTLGRR61 pKa = 11.84GSSLQLTPQLLNHH74 pKa = 6.01QAGHH78 pKa = 5.01VTTGDD83 pKa = 3.47EE84 pKa = 4.49LTPSLTPSSSTPAVLPEE101 pKa = 4.01QVFSADD107 pKa = 4.22APTQTPEE114 pKa = 3.6AAAPEE119 pKa = 4.24PAPDD123 pKa = 4.03GSSLGGGHH131 pKa = 6.44SAMLLTEE138 pKa = 4.12VGGEE142 pKa = 3.94VSPEE146 pKa = 3.54IGFTTQGLSSAPEE159 pKa = 3.87FPEE162 pKa = 4.16GRR164 pKa = 11.84IEE166 pKa = 4.2GLPDD170 pKa = 3.34SSDD173 pKa = 3.12ASGSVFVPPVVVVPPTEE190 pKa = 4.13PEE192 pKa = 4.06PPVEE196 pKa = 4.03PEE198 pKa = 4.15PPVEE202 pKa = 4.22HH203 pKa = 6.71PVDD206 pKa = 3.63HH207 pKa = 6.44GVEE210 pKa = 3.95LSGGQLTLAEE220 pKa = 5.17GDD222 pKa = 4.14LSCGPVTQSGVFTVNAVDD240 pKa = 4.25GLQSLVVGGLAVISGGVVAGFPLSTQTPLGNTFSVTAYY278 pKa = 10.47DD279 pKa = 3.72PATGEE284 pKa = 3.83VSYY287 pKa = 11.07SYY289 pKa = 9.63TLQGAIEE296 pKa = 4.29HH297 pKa = 6.52PVGEE301 pKa = 4.5GANTLGEE308 pKa = 4.42SFTVVARR315 pKa = 11.84DD316 pKa = 3.52TDD318 pKa = 3.73GDD320 pKa = 4.0QADD323 pKa = 3.76STLDD327 pKa = 3.25INIVDD332 pKa = 4.72DD333 pKa = 4.24VPEE336 pKa = 3.87AHH338 pKa = 7.04CIEE341 pKa = 4.47VTLSADD347 pKa = 3.81PDD349 pKa = 4.17CPGAPLPGASGSLLQGGTFGADD371 pKa = 2.48GGFIHH376 pKa = 7.13SISLNGTTYY385 pKa = 10.73SYY387 pKa = 11.4DD388 pKa = 3.59PEE390 pKa = 4.77TGCSTGSCEE399 pKa = 4.16GTGVFDD405 pKa = 4.04PATYY409 pKa = 10.71SLTVTSEE416 pKa = 3.97GGGTLVVNMLNGDD429 pKa = 4.24FSYY432 pKa = 10.84TPSTNPACEE441 pKa = 3.83PLTEE445 pKa = 4.6NIHH448 pKa = 6.05YY449 pKa = 9.96VISDD453 pKa = 3.56NDD455 pKa = 3.49GDD457 pKa = 4.35LAGSDD462 pKa = 3.87LQIHH466 pKa = 6.0VPAILPPEE474 pKa = 4.68GPTAVVDD481 pKa = 3.86NVITNILAPCIEE493 pKa = 4.46VPGVLLVTNDD503 pKa = 3.32VPGSGGPLTASPTVFHH519 pKa = 6.65TGWQVKK525 pKa = 10.63GEE527 pKa = 4.22GFSNDD532 pKa = 3.29CQKK535 pKa = 10.0TIDD538 pKa = 3.96FSGKK542 pKa = 9.3RR543 pKa = 11.84DD544 pKa = 3.42NSANQLKK551 pKa = 10.2NLEE554 pKa = 4.27RR555 pKa = 11.84SDD557 pKa = 3.98FANTGAMTALVVITGYY573 pKa = 10.9LGAFSGPGANAQDD586 pKa = 4.31LYY588 pKa = 11.35SVNLMAGEE596 pKa = 4.32SVAVDD601 pKa = 4.23LKK603 pKa = 11.49SLGDD607 pKa = 3.49QVGVFWQLDD616 pKa = 3.46GGEE619 pKa = 4.41FQPLDD624 pKa = 3.45AGGTFTATEE633 pKa = 3.89NGVYY637 pKa = 10.3RR638 pKa = 11.84LLVVNQPDD646 pKa = 3.45AGQPGPAVHH655 pKa = 5.73YY656 pKa = 9.45TLDD659 pKa = 3.7LVIDD663 pKa = 3.9YY664 pKa = 10.28SAADD668 pKa = 3.49TTPNHH673 pKa = 6.07EE674 pKa = 4.1GAYY677 pKa = 8.45TVNDD681 pKa = 3.52GHH683 pKa = 6.62GGSDD687 pKa = 3.09SAAVNISYY695 pKa = 10.58QEE697 pKa = 4.24GCTLLGTTGDD707 pKa = 3.95DD708 pKa = 3.55VLLGGAQDD716 pKa = 4.41DD717 pKa = 4.51SLHH720 pKa = 6.65GGDD723 pKa = 4.84GDD725 pKa = 4.46DD726 pKa = 4.09VLSGGAGNNDD736 pKa = 3.21LYY738 pKa = 11.37GDD740 pKa = 3.93NGNDD744 pKa = 3.37VLFSGSGNDD753 pKa = 3.67VLDD756 pKa = 4.39GGAGNNTASYY766 pKa = 10.87ALADD770 pKa = 3.6SGVTVSTAQLGPQHH784 pKa = 6.5TGGAGTDD791 pKa = 3.39TLINIQNLTGSDD803 pKa = 3.95YY804 pKa = 11.33DD805 pKa = 3.61DD806 pKa = 5.18HH807 pKa = 6.88LTGDD811 pKa = 3.74YY812 pKa = 8.44EE813 pKa = 5.1ANVINGGLGNDD824 pKa = 3.62VLVGGLGNDD833 pKa = 3.71TLTGGAGHH841 pKa = 7.22DD842 pKa = 3.69TFKK845 pKa = 10.78WLAGDD850 pKa = 3.84SGHH853 pKa = 7.49DD854 pKa = 3.6LVTDD858 pKa = 4.35FSLGADD864 pKa = 3.41TLDD867 pKa = 4.64LSKK870 pKa = 10.89LLQGFSVGTDD880 pKa = 3.14SLDD883 pKa = 3.66DD884 pKa = 3.86FLHH887 pKa = 6.36FKK889 pKa = 9.13VTGSGADD896 pKa = 3.33LVSSIEE902 pKa = 3.98IGSGGSSQTIDD913 pKa = 3.25LAGVNLAQHH922 pKa = 6.34YY923 pKa = 9.9DD924 pKa = 3.5VGVGAGGIVAGADD937 pKa = 3.02AGSIISGMLGDD948 pKa = 3.91HH949 pKa = 6.4SLRR952 pKa = 11.84ADD954 pKa = 3.56VVV956 pKa = 3.47
Molecular weight: 96.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y3P3S5|A0A1Y3P3S5_9PSED TetR family transcriptional regulator OS=Pseudomonas caspiana OX=1451454 GN=AUC60_23615 PE=4 SV=1
MM1 pKa = 8.22DD2 pKa = 5.9DD3 pKa = 4.11ALIAALLTQVTFWLCMSLFIRR24 pKa = 11.84LFTFRR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 9.24GARR34 pKa = 11.84FRR36 pKa = 11.84RR37 pKa = 11.84DD38 pKa = 3.31MSCLAWLMMASAGAAMVYY56 pKa = 9.64IGKK59 pKa = 10.33GMLIVQVTAWPLVLILAVFVGSVYY83 pKa = 10.45QSSGNLARR91 pKa = 11.84VWKK94 pKa = 10.63VGG96 pKa = 3.19
MM1 pKa = 8.22DD2 pKa = 5.9DD3 pKa = 4.11ALIAALLTQVTFWLCMSLFIRR24 pKa = 11.84LFTFRR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 9.24GARR34 pKa = 11.84FRR36 pKa = 11.84RR37 pKa = 11.84DD38 pKa = 3.31MSCLAWLMMASAGAAMVYY56 pKa = 9.64IGKK59 pKa = 10.33GMLIVQVTAWPLVLILAVFVGSVYY83 pKa = 10.45QSSGNLARR91 pKa = 11.84VWKK94 pKa = 10.63VGG96 pKa = 3.19
Molecular weight: 10.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1716129 |
36 |
5915 |
325.0 |
35.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.631 ± 0.038 | 0.937 ± 0.011 |
5.411 ± 0.026 | 5.702 ± 0.026 |
3.707 ± 0.021 | 7.74 ± 0.034 |
2.183 ± 0.016 | 5.114 ± 0.028 |
3.757 ± 0.031 | 11.471 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.017 | 3.219 ± 0.023 |
4.764 ± 0.024 | 4.466 ± 0.027 |
6.11 ± 0.029 | 6.185 ± 0.025 |
5.058 ± 0.025 | 7.178 ± 0.027 |
1.407 ± 0.015 | 2.54 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
