
Cinara cedri
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Paraneoptera; Hemiptera; Sternorrhyncha; Aphidomorpha; Aphidoidea;
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
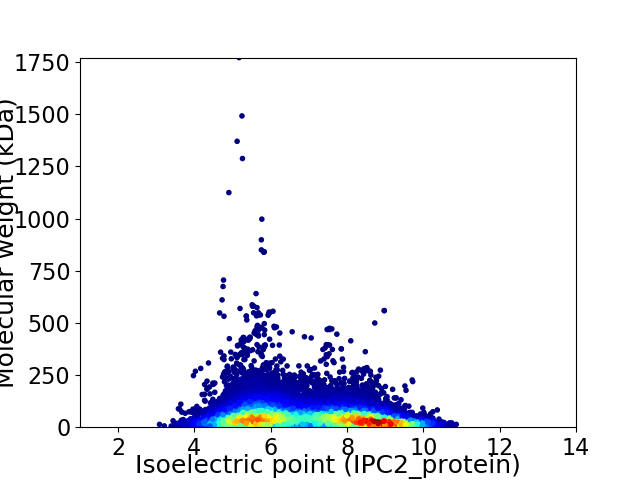
Virtual 2D-PAGE plot for 22140 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5E4M159|A0A5E4M159_9HEMI Isoform of A0A5E4M372 Uncharacterized protein OS=Cinara cedri OX=506608 GN=CINCED_3A021463 PE=4 SV=1
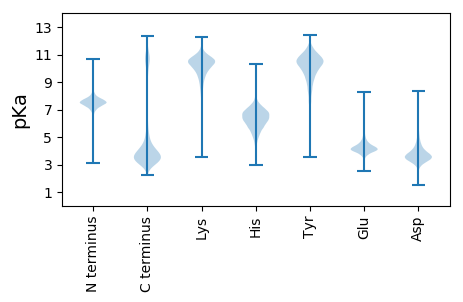
MM1 pKa = 7.93RR2 pKa = 11.84LFLYY6 pKa = 9.78TLLVSTLAISKK17 pKa = 7.37TVRR20 pKa = 11.84TDD22 pKa = 2.91NPQPSDD28 pKa = 3.43QTNLEE33 pKa = 4.19LSVSNDD39 pKa = 2.97QSNSYY44 pKa = 10.41QNQNSLKK51 pKa = 10.65ADD53 pKa = 3.37VTNDD57 pKa = 3.22GKK59 pKa = 10.26HH60 pKa = 5.97ASLDD64 pKa = 3.55ASSQTVQNSQSKK76 pKa = 10.82NEE78 pKa = 3.72LHH80 pKa = 7.09LNIEE84 pKa = 4.21QSDD87 pKa = 4.0TDD89 pKa = 3.91SDD91 pKa = 4.63PNDD94 pKa = 3.52STISNSAMTNEE105 pKa = 5.09NINTLQQQSYY115 pKa = 9.19NFKK118 pKa = 10.38EE119 pKa = 4.49SNSLLLNMQNNNIDD133 pKa = 3.53NDD135 pKa = 4.08NNNSPDD141 pKa = 3.42SGTDD145 pKa = 3.32DD146 pKa = 5.18LEE148 pKa = 5.02SNNPTGQNFGLISPQPDD165 pKa = 3.45GASPQILGSGIQTNSPTQGQNIMSLSGTDD194 pKa = 3.34PNSINNADD202 pKa = 4.04AYY204 pKa = 10.81GPTQPRR210 pKa = 11.84DD211 pKa = 3.32SSSPQGSPINYY222 pKa = 9.13NDD224 pKa = 4.54DD225 pKa = 4.58NIPTAGGSSNGNTPPSSGGTTSPDD249 pKa = 2.9SSQIGSGSLPISTSPGGQNGLNPSLFDD276 pKa = 4.4PNSPNDD282 pKa = 3.59AGTFGPSQFPDD293 pKa = 3.13SSSLQGSPNNNNNDD307 pKa = 4.09NIPPAGGISNGNTPPSSGGPASPDD331 pKa = 3.16SPQIGSGSLPISTSPGGQNGLNPSLFDD358 pKa = 4.4PNSPNDD364 pKa = 3.59AGTFGPSQFPDD375 pKa = 3.13SSSLQGSPNNNNNDD389 pKa = 4.09NIPPAGGSSNGNTPPSSGGPASPSYY414 pKa = 9.33PQFGIGLVPLSYY426 pKa = 10.61LPSGQNGVNPSLFDD440 pKa = 4.33PNSINNAGAFGPSQPPDD457 pKa = 3.22SSSPQGSPNNNNNDD471 pKa = 4.09NIPPAGGSSNGNTPPSSGGPASPDD495 pKa = 3.16SPQIGSGSIPISTSPGGQNGLNPSLFDD522 pKa = 4.4PNSPNDD528 pKa = 3.75AGSNDD533 pKa = 3.68AGSFGPSQFPDD544 pKa = 3.13SSSLQGSPNNNNNDD558 pKa = 4.09NIPPAGGLSNGNSPPSSEE576 pKa = 4.09GSSTVTNLYY585 pKa = 8.82TKK587 pKa = 10.52SSEE590 pKa = 4.18LSNSEE595 pKa = 4.02YY596 pKa = 10.8FNGGVSHH603 pKa = 6.68SEE605 pKa = 3.6SSQFEE610 pKa = 3.93EE611 pKa = 4.94SISITSDD618 pKa = 2.94STEE621 pKa = 4.23MEE623 pKa = 4.05TQQYY627 pKa = 9.6VLNSYY632 pKa = 10.96SSGKK636 pKa = 9.69FICDD640 pKa = 3.34GYY642 pKa = 11.28GKK644 pKa = 10.42IIGVVFNHH652 pKa = 6.4SADD655 pKa = 3.56SGQGLDD661 pKa = 5.1TIDD664 pKa = 5.27LDD666 pKa = 4.22TNSVIWEE673 pKa = 4.75DD674 pKa = 3.53INMLGLDD681 pKa = 3.78IYY683 pKa = 10.97QFSGNAGNYY692 pKa = 10.02VGDD695 pKa = 4.21KK696 pKa = 10.74ADD698 pKa = 3.44NAYY701 pKa = 9.36TSSKK705 pKa = 10.91NSLGDD710 pKa = 3.25AANYY714 pKa = 10.39VGDD717 pKa = 4.29KK718 pKa = 10.94ADD720 pKa = 3.58NAYY723 pKa = 7.7TASKK727 pKa = 10.88NSLGDD732 pKa = 3.26AANYY736 pKa = 10.39VGDD739 pKa = 4.29KK740 pKa = 10.83ADD742 pKa = 3.44NAYY745 pKa = 9.36TSSKK749 pKa = 10.91NSLGDD754 pKa = 3.49AANHH758 pKa = 6.35VGDD761 pKa = 4.44IADD764 pKa = 3.64NAYY767 pKa = 9.31TSSKK771 pKa = 10.91NSLGDD776 pKa = 3.25AANYY780 pKa = 10.39VGDD783 pKa = 3.83KK784 pKa = 10.93AGAVNDD790 pKa = 4.0ASKK793 pKa = 10.74EE794 pKa = 4.04AGNSIHH800 pKa = 7.01SATNDD805 pKa = 3.62YY806 pKa = 11.37GHH808 pKa = 7.5DD809 pKa = 4.03LYY811 pKa = 11.44DD812 pKa = 3.96AGHH815 pKa = 6.21NFKK818 pKa = 10.84EE819 pKa = 4.25NAFGSSDD826 pKa = 3.38GDD828 pKa = 3.59VTNNVV833 pKa = 3.18
MM1 pKa = 7.93RR2 pKa = 11.84LFLYY6 pKa = 9.78TLLVSTLAISKK17 pKa = 7.37TVRR20 pKa = 11.84TDD22 pKa = 2.91NPQPSDD28 pKa = 3.43QTNLEE33 pKa = 4.19LSVSNDD39 pKa = 2.97QSNSYY44 pKa = 10.41QNQNSLKK51 pKa = 10.65ADD53 pKa = 3.37VTNDD57 pKa = 3.22GKK59 pKa = 10.26HH60 pKa = 5.97ASLDD64 pKa = 3.55ASSQTVQNSQSKK76 pKa = 10.82NEE78 pKa = 3.72LHH80 pKa = 7.09LNIEE84 pKa = 4.21QSDD87 pKa = 4.0TDD89 pKa = 3.91SDD91 pKa = 4.63PNDD94 pKa = 3.52STISNSAMTNEE105 pKa = 5.09NINTLQQQSYY115 pKa = 9.19NFKK118 pKa = 10.38EE119 pKa = 4.49SNSLLLNMQNNNIDD133 pKa = 3.53NDD135 pKa = 4.08NNNSPDD141 pKa = 3.42SGTDD145 pKa = 3.32DD146 pKa = 5.18LEE148 pKa = 5.02SNNPTGQNFGLISPQPDD165 pKa = 3.45GASPQILGSGIQTNSPTQGQNIMSLSGTDD194 pKa = 3.34PNSINNADD202 pKa = 4.04AYY204 pKa = 10.81GPTQPRR210 pKa = 11.84DD211 pKa = 3.32SSSPQGSPINYY222 pKa = 9.13NDD224 pKa = 4.54DD225 pKa = 4.58NIPTAGGSSNGNTPPSSGGTTSPDD249 pKa = 2.9SSQIGSGSLPISTSPGGQNGLNPSLFDD276 pKa = 4.4PNSPNDD282 pKa = 3.59AGTFGPSQFPDD293 pKa = 3.13SSSLQGSPNNNNNDD307 pKa = 4.09NIPPAGGISNGNTPPSSGGPASPDD331 pKa = 3.16SPQIGSGSLPISTSPGGQNGLNPSLFDD358 pKa = 4.4PNSPNDD364 pKa = 3.59AGTFGPSQFPDD375 pKa = 3.13SSSLQGSPNNNNNDD389 pKa = 4.09NIPPAGGSSNGNTPPSSGGPASPSYY414 pKa = 9.33PQFGIGLVPLSYY426 pKa = 10.61LPSGQNGVNPSLFDD440 pKa = 4.33PNSINNAGAFGPSQPPDD457 pKa = 3.22SSSPQGSPNNNNNDD471 pKa = 4.09NIPPAGGSSNGNTPPSSGGPASPDD495 pKa = 3.16SPQIGSGSIPISTSPGGQNGLNPSLFDD522 pKa = 4.4PNSPNDD528 pKa = 3.75AGSNDD533 pKa = 3.68AGSFGPSQFPDD544 pKa = 3.13SSSLQGSPNNNNNDD558 pKa = 4.09NIPPAGGLSNGNSPPSSEE576 pKa = 4.09GSSTVTNLYY585 pKa = 8.82TKK587 pKa = 10.52SSEE590 pKa = 4.18LSNSEE595 pKa = 4.02YY596 pKa = 10.8FNGGVSHH603 pKa = 6.68SEE605 pKa = 3.6SSQFEE610 pKa = 3.93EE611 pKa = 4.94SISITSDD618 pKa = 2.94STEE621 pKa = 4.23MEE623 pKa = 4.05TQQYY627 pKa = 9.6VLNSYY632 pKa = 10.96SSGKK636 pKa = 9.69FICDD640 pKa = 3.34GYY642 pKa = 11.28GKK644 pKa = 10.42IIGVVFNHH652 pKa = 6.4SADD655 pKa = 3.56SGQGLDD661 pKa = 5.1TIDD664 pKa = 5.27LDD666 pKa = 4.22TNSVIWEE673 pKa = 4.75DD674 pKa = 3.53INMLGLDD681 pKa = 3.78IYY683 pKa = 10.97QFSGNAGNYY692 pKa = 10.02VGDD695 pKa = 4.21KK696 pKa = 10.74ADD698 pKa = 3.44NAYY701 pKa = 9.36TSSKK705 pKa = 10.91NSLGDD710 pKa = 3.25AANYY714 pKa = 10.39VGDD717 pKa = 4.29KK718 pKa = 10.94ADD720 pKa = 3.58NAYY723 pKa = 7.7TASKK727 pKa = 10.88NSLGDD732 pKa = 3.26AANYY736 pKa = 10.39VGDD739 pKa = 4.29KK740 pKa = 10.83ADD742 pKa = 3.44NAYY745 pKa = 9.36TSSKK749 pKa = 10.91NSLGDD754 pKa = 3.49AANHH758 pKa = 6.35VGDD761 pKa = 4.44IADD764 pKa = 3.64NAYY767 pKa = 9.31TSSKK771 pKa = 10.91NSLGDD776 pKa = 3.25AANYY780 pKa = 10.39VGDD783 pKa = 3.83KK784 pKa = 10.93AGAVNDD790 pKa = 4.0ASKK793 pKa = 10.74EE794 pKa = 4.04AGNSIHH800 pKa = 7.01SATNDD805 pKa = 3.62YY806 pKa = 11.37GHH808 pKa = 7.5DD809 pKa = 4.03LYY811 pKa = 11.44DD812 pKa = 3.96AGHH815 pKa = 6.21NFKK818 pKa = 10.84EE819 pKa = 4.25NAFGSSDD826 pKa = 3.38GDD828 pKa = 3.59VTNNVV833 pKa = 3.18
Molecular weight: 85.05 kDa
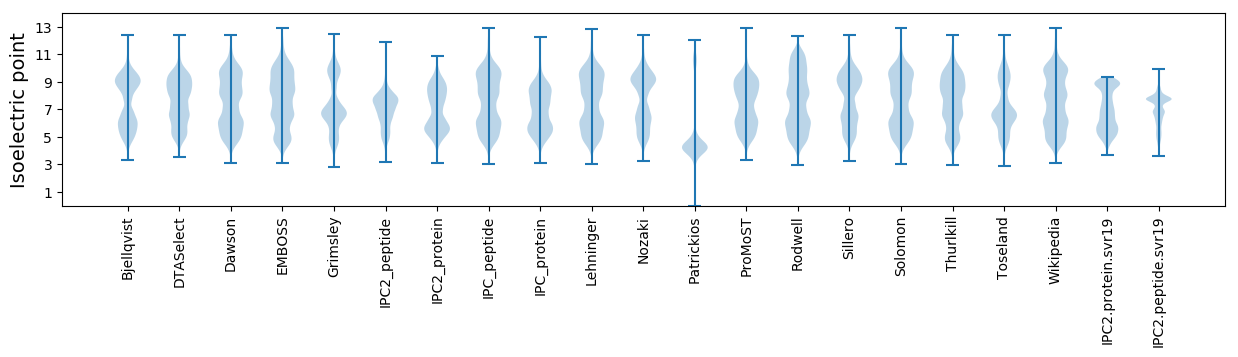
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5E4NGA4|A0A5E4NGA4_9HEMI 39S ribosomal protein L28 mitochondrial OS=Cinara cedri OX=506608 GN=CINCED_3A006303 PE=3 SV=1
MM1 pKa = 7.65GGQKK5 pKa = 9.56KK6 pKa = 9.38AAAEE10 pKa = 3.99TLSLGGGGPRR20 pKa = 11.84TPGVYY25 pKa = 9.15TRR27 pKa = 11.84AAFFNSAPGQTHH39 pKa = 5.34CRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84TTRR49 pKa = 11.84CRR51 pKa = 11.84FVAPRR56 pKa = 11.84TTPPPVVYY64 pKa = 9.15PAFPLRR70 pKa = 11.84SIRR73 pKa = 11.84SPPPPPPPPKK83 pKa = 10.35SPPEE87 pKa = 4.17HH88 pKa = 6.18VCSTTKK94 pKa = 10.13GHH96 pKa = 6.93CASQPLAPGKK106 pKa = 9.86WNDD109 pKa = 2.71IFFGIFRR116 pKa = 11.84DD117 pKa = 3.88STHH120 pKa = 7.43DD121 pKa = 3.38EE122 pKa = 4.51LYY124 pKa = 10.25TLGFRR129 pKa = 11.84EE130 pKa = 4.1RR131 pKa = 11.84PLDD134 pKa = 3.73EE135 pKa = 5.2KK136 pKa = 11.11SRR138 pKa = 11.84CDD140 pKa = 3.14RR141 pKa = 11.84TVPSGDD147 pKa = 3.34VFSS150 pKa = 4.53
MM1 pKa = 7.65GGQKK5 pKa = 9.56KK6 pKa = 9.38AAAEE10 pKa = 3.99TLSLGGGGPRR20 pKa = 11.84TPGVYY25 pKa = 9.15TRR27 pKa = 11.84AAFFNSAPGQTHH39 pKa = 5.34CRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84TTRR49 pKa = 11.84CRR51 pKa = 11.84FVAPRR56 pKa = 11.84TTPPPVVYY64 pKa = 9.15PAFPLRR70 pKa = 11.84SIRR73 pKa = 11.84SPPPPPPPPKK83 pKa = 10.35SPPEE87 pKa = 4.17HH88 pKa = 6.18VCSTTKK94 pKa = 10.13GHH96 pKa = 6.93CASQPLAPGKK106 pKa = 9.86WNDD109 pKa = 2.71IFFGIFRR116 pKa = 11.84DD117 pKa = 3.88STHH120 pKa = 7.43DD121 pKa = 3.38EE122 pKa = 4.51LYY124 pKa = 10.25TLGFRR129 pKa = 11.84EE130 pKa = 4.1RR131 pKa = 11.84PLDD134 pKa = 3.73EE135 pKa = 5.2KK136 pKa = 11.11SRR138 pKa = 11.84CDD140 pKa = 3.14RR141 pKa = 11.84TVPSGDD147 pKa = 3.34VFSS150 pKa = 4.53
Molecular weight: 16.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
10829743 |
38 |
16606 |
489.1 |
55.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.286 ± 0.018 | 2.051 ± 0.034 |
5.558 ± 0.013 | 6.231 ± 0.019 |
3.914 ± 0.015 | 5.058 ± 0.023 |
2.426 ± 0.008 | 6.562 ± 0.019 |
7.139 ± 0.023 | 8.952 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.379 ± 0.008 | 6.232 ± 0.018 |
4.742 ± 0.026 | 4.167 ± 0.021 |
4.637 ± 0.017 | 8.209 ± 0.021 |
5.957 ± 0.013 | 6.028 ± 0.014 |
1.055 ± 0.007 | 3.398 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
