
Leucobacter sp. HDW9A
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Leucobacter
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
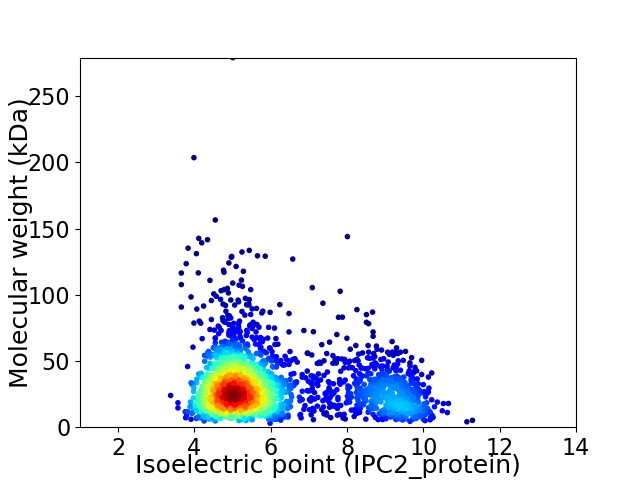
Virtual 2D-PAGE plot for 2376 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G8FPR9|A0A6G8FPR9_9MICO DUF2200 domain-containing protein OS=Leucobacter sp. HDW9A OX=2714933 GN=G7066_06465 PE=4 SV=1
MM1 pKa = 6.93TAVLVDD7 pKa = 3.48EE8 pKa = 5.37FGNAVSGEE16 pKa = 4.14EE17 pKa = 3.94TALLATTADD26 pKa = 3.9GLGSGGITAFTEE38 pKa = 4.52SVVPGTYY45 pKa = 8.78TAAVTSSVAGNKK57 pKa = 8.25TLAVALSGEE66 pKa = 4.29PVLPAGNTIARR77 pKa = 11.84FVAGGVDD84 pKa = 3.32TGNPGTTYY92 pKa = 10.72AVSGGQQTVGTGAHH106 pKa = 7.07TITVTLVDD114 pKa = 3.66AEE116 pKa = 4.59GNPVPAQAAGISADD130 pKa = 3.64TGDD133 pKa = 4.37DD134 pKa = 3.9LGDD137 pKa = 3.65GSISDD142 pKa = 3.81FTEE145 pKa = 3.74TGTAGTYY152 pKa = 6.87TASVTSTVSGPKK164 pKa = 9.66TITVLYY170 pKa = 9.17GANPVTLAGNDD181 pKa = 3.26TALFVPGAIDD191 pKa = 3.43FTNANTNYY199 pKa = 9.14TVSTGTRR206 pKa = 11.84PVGAGEE212 pKa = 4.09HH213 pKa = 5.16TVTVILSDD221 pKa = 3.46EE222 pKa = 4.43FGNPVSGQQAAIVPATADD240 pKa = 3.25ALGTGEE246 pKa = 4.76FSAFTEE252 pKa = 4.26TATAGTYY259 pKa = 7.66EE260 pKa = 4.11ATVTSSVSGVKK271 pKa = 10.38AITVSVGVNSVRR283 pKa = 11.84LSGNGTAAFVSGDD296 pKa = 3.14VDD298 pKa = 4.7LEE300 pKa = 4.14NSEE303 pKa = 4.26TNYY306 pKa = 10.19VVTTGDD312 pKa = 3.24VSVDD316 pKa = 3.49GGSHH320 pKa = 4.95SVIVTLADD328 pKa = 3.49EE329 pKa = 4.93LNNPVTGQSADD340 pKa = 3.81LLASTINSLGSGTITGFTEE359 pKa = 3.85TAIPGEE365 pKa = 4.21YY366 pKa = 9.63QATITSSIAGNKK378 pKa = 9.89DD379 pKa = 2.69ITVSLGGEE387 pKa = 4.04PVTAAGNTAASFVAGGVDD405 pKa = 3.39VGNSGTRR412 pKa = 11.84YY413 pKa = 9.67SVSAGNQTVSTGSHH427 pKa = 4.85TVTVTLADD435 pKa = 3.89AEE437 pKa = 4.65DD438 pKa = 3.9NAVSGQAAGLSATTAADD455 pKa = 3.68LGTGEE460 pKa = 4.7IGTFTEE466 pKa = 4.25TATAGTYY473 pKa = 9.1QATITSTVSGPKK485 pKa = 9.9PITVTYY491 pKa = 9.97GGSAVTASGNTTAVFVAGEE510 pKa = 3.93VDD512 pKa = 4.28LNNGSTRR519 pKa = 11.84YY520 pKa = 10.03SVTTGDD526 pKa = 3.46QVVGSGQHH534 pKa = 4.88TVTVTLRR541 pKa = 11.84DD542 pKa = 3.64EE543 pKa = 4.76FNNPVTNQASNLVADD558 pKa = 4.44SAQNLGVGTISPFTEE573 pKa = 4.24TSDD576 pKa = 3.64GVYY579 pKa = 9.78EE580 pKa = 4.44ASLSSTVSGSKK591 pKa = 10.57NITVALGLASVPLSGNGVASFVAGSVDD618 pKa = 4.11LNDD621 pKa = 3.56SGTVYY626 pKa = 10.65SVSTGEE632 pKa = 4.09QQVGTGLHH640 pKa = 6.62TITVTLLDD648 pKa = 3.6VFGNGVPGQQALIDD662 pKa = 4.39ANTSAVIGTGSFSAFAPTATPGIYY686 pKa = 9.0TATVSSSISGSKK698 pKa = 10.12PIDD701 pKa = 3.41VSFEE705 pKa = 3.79GAAVDD710 pKa = 4.57ASGNTDD716 pKa = 3.21ALFAAGGVDD725 pKa = 3.53PANPASNYY733 pKa = 10.03AVSTGDD739 pKa = 3.72EE740 pKa = 4.08IVGTGSHH747 pKa = 5.46TVTVALADD755 pKa = 3.67ALGNPVPGQAAQLAATTSGDD775 pKa = 3.46LGTGAFTGFVEE786 pKa = 4.57SASVGIYY793 pKa = 8.59EE794 pKa = 4.43ATMTSTVSGSKK805 pKa = 10.14PVTVTYY811 pKa = 10.92NSAPITLAGNGSASFISGDD830 pKa = 3.49TDD832 pKa = 3.52LTSPATSYY840 pKa = 11.12SVSSGNASVEE850 pKa = 4.43GGSHH854 pKa = 7.19LVTITLADD862 pKa = 3.64SFANPVPGKK871 pKa = 10.43AVDD874 pKa = 4.44LVASTTDD881 pKa = 3.53DD882 pKa = 4.16LGSGTITAVTEE893 pKa = 4.24SLTTPGTYY901 pKa = 9.65QATVTSSVAGGKK913 pKa = 10.07AIAVALGGDD922 pKa = 3.61SVTLNGNGTAQFIAGGPDD940 pKa = 3.14TDD942 pKa = 3.59NAGTVFEE949 pKa = 4.52VTEE952 pKa = 4.65GPQTVGAGSHH962 pKa = 4.53TVRR965 pKa = 11.84VNLRR969 pKa = 11.84DD970 pKa = 3.42SSGNPVSGEE979 pKa = 3.68AAGINANTVDD989 pKa = 3.85GLGSGTISSFTEE1001 pKa = 3.77TGTPGLYY1008 pKa = 9.27EE1009 pKa = 4.23ASVTSTLIGGKK1020 pKa = 9.96SITVNYY1026 pKa = 8.47GASAITAQGNTVALFVAAAVDD1047 pKa = 3.86PGSPGTNYY1055 pKa = 10.61SVTSGNQQVGTGQHH1069 pKa = 5.09TVTVTLADD1077 pKa = 3.28AFGNRR1082 pKa = 11.84VSGQSALLDD1091 pKa = 3.77AATTDD1096 pKa = 3.67SLGSGTIGGFVEE1108 pKa = 4.42TGTTGVYY1115 pKa = 9.84LAPVTSTLAGSKK1127 pKa = 10.4AVTASYY1133 pKa = 11.26NSLPITLSGNGNAVFVAGGVDD1154 pKa = 3.35VGNAATSYY1162 pKa = 9.42TVSSGDD1168 pKa = 3.36ASVSSGSHH1176 pKa = 6.57TITMTLADD1184 pKa = 3.86AFGNPVPGMAPRR1196 pKa = 11.84VTATTAANLGSGTITGVTEE1215 pKa = 3.63QATPGTYY1222 pKa = 9.68QATVTSSIAGNKK1234 pKa = 9.97DD1235 pKa = 2.45ITALFDD1241 pKa = 3.98AAPLTLSGNGIARR1254 pKa = 11.84FVAGEE1259 pKa = 4.73LILQTRR1265 pKa = 11.84APLSPP1270 pKa = 3.86
MM1 pKa = 6.93TAVLVDD7 pKa = 3.48EE8 pKa = 5.37FGNAVSGEE16 pKa = 4.14EE17 pKa = 3.94TALLATTADD26 pKa = 3.9GLGSGGITAFTEE38 pKa = 4.52SVVPGTYY45 pKa = 8.78TAAVTSSVAGNKK57 pKa = 8.25TLAVALSGEE66 pKa = 4.29PVLPAGNTIARR77 pKa = 11.84FVAGGVDD84 pKa = 3.32TGNPGTTYY92 pKa = 10.72AVSGGQQTVGTGAHH106 pKa = 7.07TITVTLVDD114 pKa = 3.66AEE116 pKa = 4.59GNPVPAQAAGISADD130 pKa = 3.64TGDD133 pKa = 4.37DD134 pKa = 3.9LGDD137 pKa = 3.65GSISDD142 pKa = 3.81FTEE145 pKa = 3.74TGTAGTYY152 pKa = 6.87TASVTSTVSGPKK164 pKa = 9.66TITVLYY170 pKa = 9.17GANPVTLAGNDD181 pKa = 3.26TALFVPGAIDD191 pKa = 3.43FTNANTNYY199 pKa = 9.14TVSTGTRR206 pKa = 11.84PVGAGEE212 pKa = 4.09HH213 pKa = 5.16TVTVILSDD221 pKa = 3.46EE222 pKa = 4.43FGNPVSGQQAAIVPATADD240 pKa = 3.25ALGTGEE246 pKa = 4.76FSAFTEE252 pKa = 4.26TATAGTYY259 pKa = 7.66EE260 pKa = 4.11ATVTSSVSGVKK271 pKa = 10.38AITVSVGVNSVRR283 pKa = 11.84LSGNGTAAFVSGDD296 pKa = 3.14VDD298 pKa = 4.7LEE300 pKa = 4.14NSEE303 pKa = 4.26TNYY306 pKa = 10.19VVTTGDD312 pKa = 3.24VSVDD316 pKa = 3.49GGSHH320 pKa = 4.95SVIVTLADD328 pKa = 3.49EE329 pKa = 4.93LNNPVTGQSADD340 pKa = 3.81LLASTINSLGSGTITGFTEE359 pKa = 3.85TAIPGEE365 pKa = 4.21YY366 pKa = 9.63QATITSSIAGNKK378 pKa = 9.89DD379 pKa = 2.69ITVSLGGEE387 pKa = 4.04PVTAAGNTAASFVAGGVDD405 pKa = 3.39VGNSGTRR412 pKa = 11.84YY413 pKa = 9.67SVSAGNQTVSTGSHH427 pKa = 4.85TVTVTLADD435 pKa = 3.89AEE437 pKa = 4.65DD438 pKa = 3.9NAVSGQAAGLSATTAADD455 pKa = 3.68LGTGEE460 pKa = 4.7IGTFTEE466 pKa = 4.25TATAGTYY473 pKa = 9.1QATITSTVSGPKK485 pKa = 9.9PITVTYY491 pKa = 9.97GGSAVTASGNTTAVFVAGEE510 pKa = 3.93VDD512 pKa = 4.28LNNGSTRR519 pKa = 11.84YY520 pKa = 10.03SVTTGDD526 pKa = 3.46QVVGSGQHH534 pKa = 4.88TVTVTLRR541 pKa = 11.84DD542 pKa = 3.64EE543 pKa = 4.76FNNPVTNQASNLVADD558 pKa = 4.44SAQNLGVGTISPFTEE573 pKa = 4.24TSDD576 pKa = 3.64GVYY579 pKa = 9.78EE580 pKa = 4.44ASLSSTVSGSKK591 pKa = 10.57NITVALGLASVPLSGNGVASFVAGSVDD618 pKa = 4.11LNDD621 pKa = 3.56SGTVYY626 pKa = 10.65SVSTGEE632 pKa = 4.09QQVGTGLHH640 pKa = 6.62TITVTLLDD648 pKa = 3.6VFGNGVPGQQALIDD662 pKa = 4.39ANTSAVIGTGSFSAFAPTATPGIYY686 pKa = 9.0TATVSSSISGSKK698 pKa = 10.12PIDD701 pKa = 3.41VSFEE705 pKa = 3.79GAAVDD710 pKa = 4.57ASGNTDD716 pKa = 3.21ALFAAGGVDD725 pKa = 3.53PANPASNYY733 pKa = 10.03AVSTGDD739 pKa = 3.72EE740 pKa = 4.08IVGTGSHH747 pKa = 5.46TVTVALADD755 pKa = 3.67ALGNPVPGQAAQLAATTSGDD775 pKa = 3.46LGTGAFTGFVEE786 pKa = 4.57SASVGIYY793 pKa = 8.59EE794 pKa = 4.43ATMTSTVSGSKK805 pKa = 10.14PVTVTYY811 pKa = 10.92NSAPITLAGNGSASFISGDD830 pKa = 3.49TDD832 pKa = 3.52LTSPATSYY840 pKa = 11.12SVSSGNASVEE850 pKa = 4.43GGSHH854 pKa = 7.19LVTITLADD862 pKa = 3.64SFANPVPGKK871 pKa = 10.43AVDD874 pKa = 4.44LVASTTDD881 pKa = 3.53DD882 pKa = 4.16LGSGTITAVTEE893 pKa = 4.24SLTTPGTYY901 pKa = 9.65QATVTSSVAGGKK913 pKa = 10.07AIAVALGGDD922 pKa = 3.61SVTLNGNGTAQFIAGGPDD940 pKa = 3.14TDD942 pKa = 3.59NAGTVFEE949 pKa = 4.52VTEE952 pKa = 4.65GPQTVGAGSHH962 pKa = 4.53TVRR965 pKa = 11.84VNLRR969 pKa = 11.84DD970 pKa = 3.42SSGNPVSGEE979 pKa = 3.68AAGINANTVDD989 pKa = 3.85GLGSGTISSFTEE1001 pKa = 3.77TGTPGLYY1008 pKa = 9.27EE1009 pKa = 4.23ASVTSTLIGGKK1020 pKa = 9.96SITVNYY1026 pKa = 8.47GASAITAQGNTVALFVAAAVDD1047 pKa = 3.86PGSPGTNYY1055 pKa = 10.61SVTSGNQQVGTGQHH1069 pKa = 5.09TVTVTLADD1077 pKa = 3.28AFGNRR1082 pKa = 11.84VSGQSALLDD1091 pKa = 3.77AATTDD1096 pKa = 3.67SLGSGTIGGFVEE1108 pKa = 4.42TGTTGVYY1115 pKa = 9.84LAPVTSTLAGSKK1127 pKa = 10.4AVTASYY1133 pKa = 11.26NSLPITLSGNGNAVFVAGGVDD1154 pKa = 3.35VGNAATSYY1162 pKa = 9.42TVSSGDD1168 pKa = 3.36ASVSSGSHH1176 pKa = 6.57TITMTLADD1184 pKa = 3.86AFGNPVPGMAPRR1196 pKa = 11.84VTATTAANLGSGTITGVTEE1215 pKa = 3.63QATPGTYY1222 pKa = 9.68QATVTSSIAGNKK1234 pKa = 9.97DD1235 pKa = 2.45ITALFDD1241 pKa = 3.98AAPLTLSGNGIARR1254 pKa = 11.84FVAGEE1259 pKa = 4.73LILQTRR1265 pKa = 11.84APLSPP1270 pKa = 3.86
Molecular weight: 123.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G8FMM7|A0A6G8FMM7_9MICO 3-hydroxyacyl-CoA dehydrogenase family protein OS=Leucobacter sp. HDW9A OX=2714933 GN=G7066_01125 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.5VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.45GRR40 pKa = 11.84SKK42 pKa = 10.13LTAA45 pKa = 4.04
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.5VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIVAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.45GRR40 pKa = 11.84SKK42 pKa = 10.13LTAA45 pKa = 4.04
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
718273 |
29 |
2824 |
302.3 |
32.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.096 ± 0.068 | 0.62 ± 0.014 |
5.518 ± 0.043 | 6.009 ± 0.062 |
3.317 ± 0.031 | 8.686 ± 0.055 |
1.845 ± 0.026 | 4.833 ± 0.038 |
2.778 ± 0.04 | 10.013 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.928 ± 0.021 | 2.625 ± 0.035 |
5.076 ± 0.033 | 3.242 ± 0.029 |
6.46 ± 0.074 | 6.46 ± 0.045 |
6.457 ± 0.078 | 8.631 ± 0.06 |
1.377 ± 0.021 | 2.029 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
