
Chicken anemia virus (isolate Japan 82-2) (CAV)
Taxonomy: Viruses; Anelloviridae; Gyrovirus; Chicken anemia virus
Average proteome isoelectric point is 8.49
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
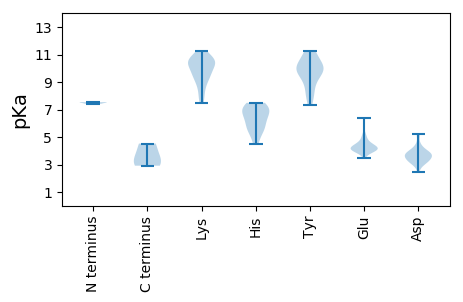
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|P54096|VP3_CAV82 Apoptin OS=Chicken anemia virus (isolate Japan 82-2) OX=73476 GN=VP3 PE=2 SV=1
MM1 pKa = 7.54HH2 pKa = 7.49GNGGQPAAGGSEE14 pKa = 4.12SALSRR19 pKa = 11.84EE20 pKa = 4.43GQPGPSGAAQGQVISNEE37 pKa = 3.83RR38 pKa = 11.84SPRR41 pKa = 11.84RR42 pKa = 11.84YY43 pKa = 8.28STRR46 pKa = 11.84TINGVQATNKK56 pKa = 7.8FTAVGNPSLQRR67 pKa = 11.84DD68 pKa = 3.84PDD70 pKa = 3.55WYY72 pKa = 9.71RR73 pKa = 11.84WNYY76 pKa = 8.1NHH78 pKa = 7.22SIAVWLRR85 pKa = 11.84EE86 pKa = 4.05CSRR89 pKa = 11.84SHH91 pKa = 7.08AKK93 pKa = 9.51ICNCGQFRR101 pKa = 11.84KK102 pKa = 9.59HH103 pKa = 5.49WFQEE107 pKa = 4.39CAGLEE112 pKa = 4.08DD113 pKa = 5.22RR114 pKa = 11.84STQASLEE121 pKa = 4.13EE122 pKa = 5.15AILRR126 pKa = 11.84PLRR129 pKa = 11.84VQGKK133 pKa = 7.45RR134 pKa = 11.84AKK136 pKa = 10.3RR137 pKa = 11.84KK138 pKa = 9.49LDD140 pKa = 3.32YY141 pKa = 10.37HH142 pKa = 6.72YY143 pKa = 11.12SQPTPNRR150 pKa = 11.84KK151 pKa = 8.91KK152 pKa = 9.96VYY154 pKa = 9.0KK155 pKa = 7.97TVRR158 pKa = 11.84WKK160 pKa = 11.07DD161 pKa = 3.51EE162 pKa = 4.08LADD165 pKa = 4.19RR166 pKa = 11.84EE167 pKa = 4.28ADD169 pKa = 3.67FTPSEE174 pKa = 4.2EE175 pKa = 4.57DD176 pKa = 3.16GGTTSSDD183 pKa = 3.08FDD185 pKa = 3.81EE186 pKa = 6.39DD187 pKa = 3.83INFDD191 pKa = 3.22IGGDD195 pKa = 3.55SGIVDD200 pKa = 4.09EE201 pKa = 4.93LLGRR205 pKa = 11.84PFTTPAPVRR214 pKa = 11.84IVV216 pKa = 2.92
MM1 pKa = 7.54HH2 pKa = 7.49GNGGQPAAGGSEE14 pKa = 4.12SALSRR19 pKa = 11.84EE20 pKa = 4.43GQPGPSGAAQGQVISNEE37 pKa = 3.83RR38 pKa = 11.84SPRR41 pKa = 11.84RR42 pKa = 11.84YY43 pKa = 8.28STRR46 pKa = 11.84TINGVQATNKK56 pKa = 7.8FTAVGNPSLQRR67 pKa = 11.84DD68 pKa = 3.84PDD70 pKa = 3.55WYY72 pKa = 9.71RR73 pKa = 11.84WNYY76 pKa = 8.1NHH78 pKa = 7.22SIAVWLRR85 pKa = 11.84EE86 pKa = 4.05CSRR89 pKa = 11.84SHH91 pKa = 7.08AKK93 pKa = 9.51ICNCGQFRR101 pKa = 11.84KK102 pKa = 9.59HH103 pKa = 5.49WFQEE107 pKa = 4.39CAGLEE112 pKa = 4.08DD113 pKa = 5.22RR114 pKa = 11.84STQASLEE121 pKa = 4.13EE122 pKa = 5.15AILRR126 pKa = 11.84PLRR129 pKa = 11.84VQGKK133 pKa = 7.45RR134 pKa = 11.84AKK136 pKa = 10.3RR137 pKa = 11.84KK138 pKa = 9.49LDD140 pKa = 3.32YY141 pKa = 10.37HH142 pKa = 6.72YY143 pKa = 11.12SQPTPNRR150 pKa = 11.84KK151 pKa = 8.91KK152 pKa = 9.96VYY154 pKa = 9.0KK155 pKa = 7.97TVRR158 pKa = 11.84WKK160 pKa = 11.07DD161 pKa = 3.51EE162 pKa = 4.08LADD165 pKa = 4.19RR166 pKa = 11.84EE167 pKa = 4.28ADD169 pKa = 3.67FTPSEE174 pKa = 4.2EE175 pKa = 4.57DD176 pKa = 3.16GGTTSSDD183 pKa = 3.08FDD185 pKa = 3.81EE186 pKa = 6.39DD187 pKa = 3.83INFDD191 pKa = 3.22IGGDD195 pKa = 3.55SGIVDD200 pKa = 4.09EE201 pKa = 4.93LLGRR205 pKa = 11.84PFTTPAPVRR214 pKa = 11.84IVV216 pKa = 2.92
Molecular weight: 24.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P54093|VP2_CAV82 Dual specificity protein phosphatase VP2 OS=Chicken anemia virus (isolate Japan 82-2) OX=73476 GN=VP2 PE=2 SV=1
MM1 pKa = 7.42EE2 pKa = 4.52RR3 pKa = 11.84RR4 pKa = 11.84ARR6 pKa = 11.84RR7 pKa = 11.84PRR9 pKa = 11.84GRR11 pKa = 11.84FYY13 pKa = 11.27AFRR16 pKa = 11.84RR17 pKa = 11.84GRR19 pKa = 11.84WNHH22 pKa = 5.65LKK24 pKa = 10.47RR25 pKa = 11.84LRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84YY30 pKa = 9.6KK31 pKa = 8.5FRR33 pKa = 11.84HH34 pKa = 5.64RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84QRR39 pKa = 11.84YY40 pKa = 7.89RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84AFRR46 pKa = 11.84KK47 pKa = 9.08AFHH50 pKa = 6.5NPRR53 pKa = 11.84PGTYY57 pKa = 9.13SVRR60 pKa = 11.84LPNPQSTMTIRR71 pKa = 11.84FQGVIFLTEE80 pKa = 3.98GLILPKK86 pKa = 10.65NSTAGGYY93 pKa = 9.63ADD95 pKa = 3.85HH96 pKa = 7.07MYY98 pKa = 10.18GARR101 pKa = 11.84VAKK104 pKa = 10.39ISVNLKK110 pKa = 9.73EE111 pKa = 4.38FLLASMNLTYY121 pKa = 10.55VSKK124 pKa = 10.97LGGPIAGEE132 pKa = 4.41LIADD136 pKa = 4.38GSKK139 pKa = 10.61SEE141 pKa = 5.18AAEE144 pKa = 3.93NWPNCCVPLDD154 pKa = 4.04NNVPSATPSAWWRR167 pKa = 11.84WALMMMQPTDD177 pKa = 2.89SCRR180 pKa = 11.84FFNHH184 pKa = 6.69PKK186 pKa = 10.56QMTLQDD192 pKa = 3.9MGRR195 pKa = 11.84MFGGWHH201 pKa = 7.18LFRR204 pKa = 11.84HH205 pKa = 5.88IEE207 pKa = 4.02TRR209 pKa = 11.84FQLLATKK216 pKa = 10.66NEE218 pKa = 4.19GSFSPVASLLSQGEE232 pKa = 4.35YY233 pKa = 8.83LTRR236 pKa = 11.84RR237 pKa = 11.84DD238 pKa = 3.7DD239 pKa = 3.65VKK241 pKa = 11.26YY242 pKa = 11.07SSDD245 pKa = 3.15HH246 pKa = 4.48QNRR249 pKa = 11.84WRR251 pKa = 11.84KK252 pKa = 9.35GGQPMTGGIAYY263 pKa = 7.3ATGKK267 pKa = 8.59MRR269 pKa = 11.84PDD271 pKa = 3.42EE272 pKa = 4.15QQYY275 pKa = 9.59PAMPPDD281 pKa = 3.92PPIITSTTAQGTQVRR296 pKa = 11.84CMNSTQAWWSWDD308 pKa = 3.04TYY310 pKa = 11.0MSFATLTALGAQWSFPPGQRR330 pKa = 11.84SVSRR334 pKa = 11.84RR335 pKa = 11.84SFNHH339 pKa = 5.11HH340 pKa = 6.41KK341 pKa = 10.76ARR343 pKa = 11.84GAGDD347 pKa = 3.45PKK349 pKa = 9.58GQRR352 pKa = 11.84WHH354 pKa = 6.22TLVPLGTEE362 pKa = 4.78TITDD366 pKa = 3.72SYY368 pKa = 10.29MGAPASEE375 pKa = 4.7LDD377 pKa = 3.69TNFFTLYY384 pKa = 9.98VAQGTNKK391 pKa = 9.19SQQYY395 pKa = 10.29KK396 pKa = 10.49FGTATYY402 pKa = 9.86ALKK405 pKa = 10.54EE406 pKa = 4.03PVMKK410 pKa = 10.25SDD412 pKa = 2.48SWAVVRR418 pKa = 11.84VQSVWQLGNRR428 pKa = 11.84QRR430 pKa = 11.84PYY432 pKa = 10.08PWDD435 pKa = 3.8VNWANSTMYY444 pKa = 10.02WGTQPP449 pKa = 4.53
MM1 pKa = 7.42EE2 pKa = 4.52RR3 pKa = 11.84RR4 pKa = 11.84ARR6 pKa = 11.84RR7 pKa = 11.84PRR9 pKa = 11.84GRR11 pKa = 11.84FYY13 pKa = 11.27AFRR16 pKa = 11.84RR17 pKa = 11.84GRR19 pKa = 11.84WNHH22 pKa = 5.65LKK24 pKa = 10.47RR25 pKa = 11.84LRR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84YY30 pKa = 9.6KK31 pKa = 8.5FRR33 pKa = 11.84HH34 pKa = 5.64RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84QRR39 pKa = 11.84YY40 pKa = 7.89RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84AFRR46 pKa = 11.84KK47 pKa = 9.08AFHH50 pKa = 6.5NPRR53 pKa = 11.84PGTYY57 pKa = 9.13SVRR60 pKa = 11.84LPNPQSTMTIRR71 pKa = 11.84FQGVIFLTEE80 pKa = 3.98GLILPKK86 pKa = 10.65NSTAGGYY93 pKa = 9.63ADD95 pKa = 3.85HH96 pKa = 7.07MYY98 pKa = 10.18GARR101 pKa = 11.84VAKK104 pKa = 10.39ISVNLKK110 pKa = 9.73EE111 pKa = 4.38FLLASMNLTYY121 pKa = 10.55VSKK124 pKa = 10.97LGGPIAGEE132 pKa = 4.41LIADD136 pKa = 4.38GSKK139 pKa = 10.61SEE141 pKa = 5.18AAEE144 pKa = 3.93NWPNCCVPLDD154 pKa = 4.04NNVPSATPSAWWRR167 pKa = 11.84WALMMMQPTDD177 pKa = 2.89SCRR180 pKa = 11.84FFNHH184 pKa = 6.69PKK186 pKa = 10.56QMTLQDD192 pKa = 3.9MGRR195 pKa = 11.84MFGGWHH201 pKa = 7.18LFRR204 pKa = 11.84HH205 pKa = 5.88IEE207 pKa = 4.02TRR209 pKa = 11.84FQLLATKK216 pKa = 10.66NEE218 pKa = 4.19GSFSPVASLLSQGEE232 pKa = 4.35YY233 pKa = 8.83LTRR236 pKa = 11.84RR237 pKa = 11.84DD238 pKa = 3.7DD239 pKa = 3.65VKK241 pKa = 11.26YY242 pKa = 11.07SSDD245 pKa = 3.15HH246 pKa = 4.48QNRR249 pKa = 11.84WRR251 pKa = 11.84KK252 pKa = 9.35GGQPMTGGIAYY263 pKa = 7.3ATGKK267 pKa = 8.59MRR269 pKa = 11.84PDD271 pKa = 3.42EE272 pKa = 4.15QQYY275 pKa = 9.59PAMPPDD281 pKa = 3.92PPIITSTTAQGTQVRR296 pKa = 11.84CMNSTQAWWSWDD308 pKa = 3.04TYY310 pKa = 11.0MSFATLTALGAQWSFPPGQRR330 pKa = 11.84SVSRR334 pKa = 11.84RR335 pKa = 11.84SFNHH339 pKa = 5.11HH340 pKa = 6.41KK341 pKa = 10.76ARR343 pKa = 11.84GAGDD347 pKa = 3.45PKK349 pKa = 9.58GQRR352 pKa = 11.84WHH354 pKa = 6.22TLVPLGTEE362 pKa = 4.78TITDD366 pKa = 3.72SYY368 pKa = 10.29MGAPASEE375 pKa = 4.7LDD377 pKa = 3.69TNFFTLYY384 pKa = 9.98VAQGTNKK391 pKa = 9.19SQQYY395 pKa = 10.29KK396 pKa = 10.49FGTATYY402 pKa = 9.86ALKK405 pKa = 10.54EE406 pKa = 4.03PVMKK410 pKa = 10.25SDD412 pKa = 2.48SWAVVRR418 pKa = 11.84VQSVWQLGNRR428 pKa = 11.84QRR430 pKa = 11.84PYY432 pKa = 10.08PWDD435 pKa = 3.8VNWANSTMYY444 pKa = 10.02WGTQPP449 pKa = 4.53
Molecular weight: 51.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
786 |
121 |
449 |
262.0 |
29.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.506 ± 0.266 | 1.654 ± 0.722 |
4.453 ± 0.943 | 4.326 ± 1.182 |
3.69 ± 0.653 | 7.506 ± 1.108 |
2.163 ± 0.365 | 3.308 ± 0.682 |
4.326 ± 0.142 | 6.234 ± 0.568 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 1.19 | 4.326 ± 0.29 |
7.379 ± 1.409 | 4.835 ± 0.873 |
10.051 ± 0.5 | 8.015 ± 0.813 |
7.634 ± 1.468 | 4.071 ± 0.464 |
2.799 ± 0.867 | 3.181 ± 0.728 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
