
Chelonia mydas (Green sea-turtle) (Chelonia agassizi)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria;
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
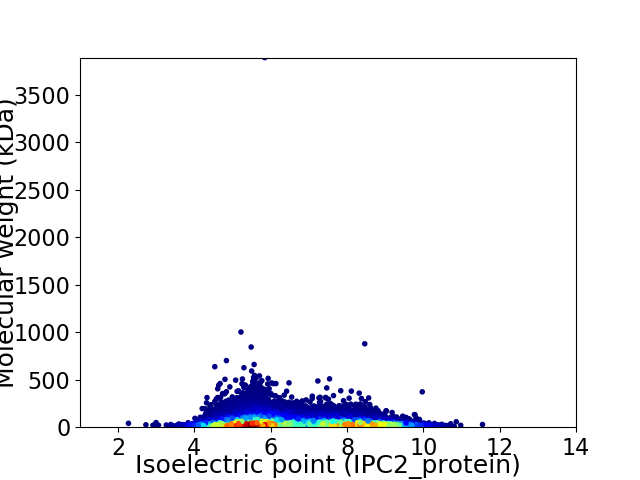
Virtual 2D-PAGE plot for 18960 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M7BXW5|M7BXW5_CHEMY EH domain-binding protein 1 OS=Chelonia mydas OX=8469 GN=UY3_09947 PE=4 SV=1
MM1 pKa = 7.32WAAVWPWTRR10 pKa = 11.84ALLLLLLFPRR20 pKa = 11.84PLAQGFLLLPGGSPAALLGLGCVLLLGLFGAALPGTLKK58 pKa = 11.05GFDD61 pKa = 3.21QTINLILDD69 pKa = 3.75EE70 pKa = 4.1SHH72 pKa = 6.5EE73 pKa = 4.5RR74 pKa = 11.84VFSSSQGVEE83 pKa = 3.89QVVLGLYY90 pKa = 9.48IVRR93 pKa = 11.84GDD95 pKa = 3.74NVAAAALDD103 pKa = 4.18NIFGTGGVIGSDD115 pKa = 2.73WCYY118 pKa = 10.72GASCTFGTDD127 pKa = 3.2DD128 pKa = 3.67GTGTSIYY135 pKa = 10.0TNIVIDD141 pKa = 4.31AGCDD145 pKa = 3.61GVDD148 pKa = 3.81EE149 pKa = 5.55DD150 pKa = 5.12PGADD154 pKa = 3.25ATPGVV159 pKa = 3.96
MM1 pKa = 7.32WAAVWPWTRR10 pKa = 11.84ALLLLLLFPRR20 pKa = 11.84PLAQGFLLLPGGSPAALLGLGCVLLLGLFGAALPGTLKK58 pKa = 11.05GFDD61 pKa = 3.21QTINLILDD69 pKa = 3.75EE70 pKa = 4.1SHH72 pKa = 6.5EE73 pKa = 4.5RR74 pKa = 11.84VFSSSQGVEE83 pKa = 3.89QVVLGLYY90 pKa = 9.48IVRR93 pKa = 11.84GDD95 pKa = 3.74NVAAAALDD103 pKa = 4.18NIFGTGGVIGSDD115 pKa = 2.73WCYY118 pKa = 10.72GASCTFGTDD127 pKa = 3.2DD128 pKa = 3.67GTGTSIYY135 pKa = 10.0TNIVIDD141 pKa = 4.31AGCDD145 pKa = 3.61GVDD148 pKa = 3.81EE149 pKa = 5.55DD150 pKa = 5.12PGADD154 pKa = 3.25ATPGVV159 pKa = 3.96
Molecular weight: 16.38 kDa
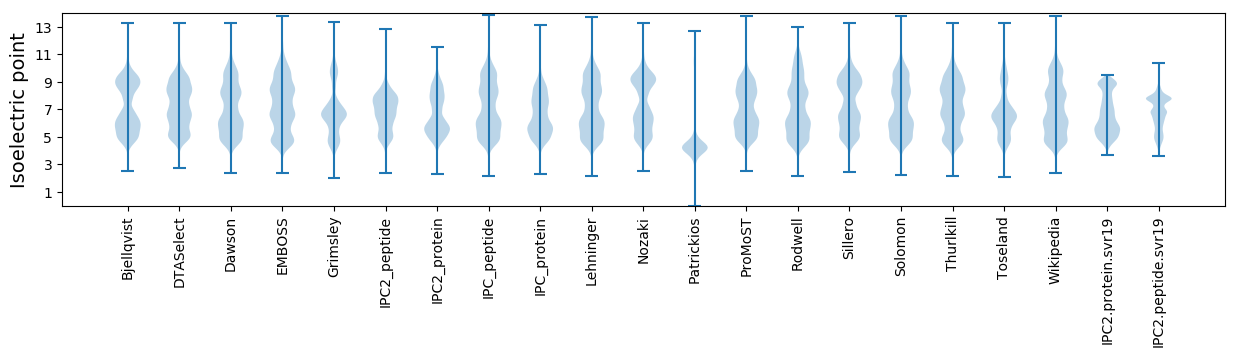
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M7ANA5|M7ANA5_CHEMY 2-phospho-D-glycerate hydro-lyase OS=Chelonia mydas OX=8469 GN=UY3_17006 PE=3 SV=1
MM1 pKa = 7.34LASTAGALRR10 pKa = 11.84PLPVMLASTAGALRR24 pKa = 11.84PLPVMLASTAGALRR38 pKa = 11.84PLPVMLASTAGALRR52 pKa = 11.84PLPVMLASTAGALRR66 pKa = 11.84PLPVMLASTAGALRR80 pKa = 11.84PLPVMLASTAGALRR94 pKa = 11.84PLPVMLASTAGALRR108 pKa = 11.84PLPVMLASTAGALRR122 pKa = 11.84PLPVMLASTAGALRR136 pKa = 11.84PLPVMLASTAGALRR150 pKa = 11.84PLPVMLASTAGALRR164 pKa = 11.84PLPVMLASTAGALRR178 pKa = 11.84PLPVMLASTAGALRR192 pKa = 11.84PLPVMLASTAGALRR206 pKa = 11.84PLPVMLASTAGALRR220 pKa = 11.84PLPVMLASTAGALRR234 pKa = 11.84PLPVMLASTAGALRR248 pKa = 11.84PLPVMLASTAGALRR262 pKa = 11.84PLPVMLASTARR273 pKa = 11.84ALRR276 pKa = 11.84PLPVMLAVVRR286 pKa = 4.34
MM1 pKa = 7.34LASTAGALRR10 pKa = 11.84PLPVMLASTAGALRR24 pKa = 11.84PLPVMLASTAGALRR38 pKa = 11.84PLPVMLASTAGALRR52 pKa = 11.84PLPVMLASTAGALRR66 pKa = 11.84PLPVMLASTAGALRR80 pKa = 11.84PLPVMLASTAGALRR94 pKa = 11.84PLPVMLASTAGALRR108 pKa = 11.84PLPVMLASTAGALRR122 pKa = 11.84PLPVMLASTAGALRR136 pKa = 11.84PLPVMLASTAGALRR150 pKa = 11.84PLPVMLASTAGALRR164 pKa = 11.84PLPVMLASTAGALRR178 pKa = 11.84PLPVMLASTAGALRR192 pKa = 11.84PLPVMLASTAGALRR206 pKa = 11.84PLPVMLASTAGALRR220 pKa = 11.84PLPVMLASTAGALRR234 pKa = 11.84PLPVMLASTAGALRR248 pKa = 11.84PLPVMLASTAGALRR262 pKa = 11.84PLPVMLASTARR273 pKa = 11.84ALRR276 pKa = 11.84PLPVMLAVVRR286 pKa = 4.34
Molecular weight: 28.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9158208 |
35 |
34967 |
483.0 |
54.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.688 ± 0.016 | 2.312 ± 0.015 |
4.901 ± 0.012 | 7.198 ± 0.025 |
3.637 ± 0.012 | 6.1 ± 0.021 |
2.599 ± 0.011 | 4.72 ± 0.015 |
6.074 ± 0.022 | 9.678 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.284 ± 0.009 | 3.914 ± 0.013 |
5.637 ± 0.021 | 4.832 ± 0.017 |
5.387 ± 0.014 | 8.431 ± 0.024 |
5.509 ± 0.016 | 6.071 ± 0.017 |
1.192 ± 0.007 | 2.802 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
