
Hubei unio douglasiae virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
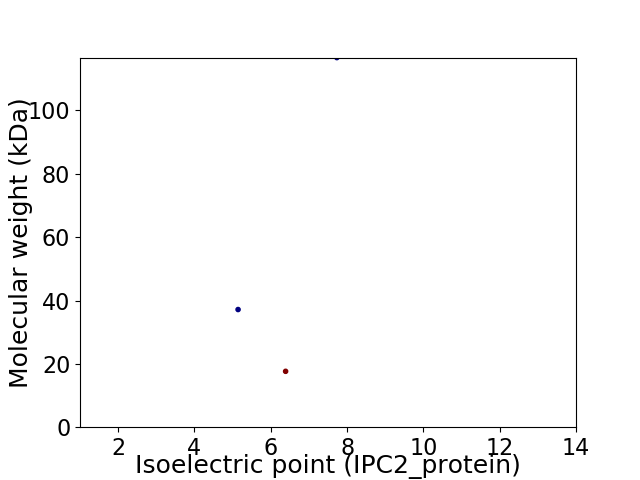
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KGU5|A0A1L3KGU5_9VIRU Capsid protein beta OS=Hubei unio douglasiae virus 1 OX=1923321 PE=4 SV=1
MM1 pKa = 7.15TLAGEE6 pKa = 4.7GFLKK10 pKa = 10.81CAFAPPDD17 pKa = 3.95FNSDD21 pKa = 3.11AGRR24 pKa = 11.84GIPDD28 pKa = 3.95DD29 pKa = 3.85FQGKK33 pKa = 8.95VLTRR37 pKa = 11.84KK38 pKa = 9.97DD39 pKa = 3.6VLTTSLSYY47 pKa = 11.02GAGQDD52 pKa = 3.47VYY54 pKa = 11.01ILVAPTPGISYY65 pKa = 8.84WYY67 pKa = 10.32ASVANGASLSGTAFRR82 pKa = 11.84PNPYY86 pKa = 9.24PGYY89 pKa = 10.07RR90 pKa = 11.84SLFGDD95 pKa = 3.59TGLEE99 pKa = 3.81RR100 pKa = 11.84ANNVTSFRR108 pKa = 11.84YY109 pKa = 10.03ASTCVGLYY117 pKa = 6.81PTSNMQQFAGSISVWKK133 pKa = 10.35FPMEE137 pKa = 5.3LINQIVGVGTPVVGQPCWVLNGTDD161 pKa = 3.3NFSGIGYY168 pKa = 9.76EE169 pKa = 4.02NYY171 pKa = 9.3TDD173 pKa = 4.07SFIKK177 pKa = 10.54GMYY180 pKa = 8.48SQSVCNEE187 pKa = 3.51PDD189 pKa = 3.15FEE191 pKa = 4.13FRR193 pKa = 11.84PIIEE197 pKa = 4.88EE198 pKa = 3.59IGAIPSGVITATQAGMFCTFATSQPAAGQLCGIVGMGTMDD238 pKa = 5.36CILIKK243 pKa = 10.7VATPVGATNSAVLKK257 pKa = 8.96TWACVEE263 pKa = 3.93YY264 pKa = 10.66RR265 pKa = 11.84PNTNSALYY273 pKa = 10.06SYY275 pKa = 10.91AHH277 pKa = 7.35DD278 pKa = 4.3SPAPDD283 pKa = 4.07LMALAAYY290 pKa = 10.19RR291 pKa = 11.84EE292 pKa = 4.13IAKK295 pKa = 9.59NVPIAVPCSQNEE307 pKa = 4.43GFWKK311 pKa = 10.25RR312 pKa = 11.84VLTILRR318 pKa = 11.84GGLSLTSKK326 pKa = 10.48IPGPIGMTATGVSGIMDD343 pKa = 4.46MMTGLATT350 pKa = 3.64
MM1 pKa = 7.15TLAGEE6 pKa = 4.7GFLKK10 pKa = 10.81CAFAPPDD17 pKa = 3.95FNSDD21 pKa = 3.11AGRR24 pKa = 11.84GIPDD28 pKa = 3.95DD29 pKa = 3.85FQGKK33 pKa = 8.95VLTRR37 pKa = 11.84KK38 pKa = 9.97DD39 pKa = 3.6VLTTSLSYY47 pKa = 11.02GAGQDD52 pKa = 3.47VYY54 pKa = 11.01ILVAPTPGISYY65 pKa = 8.84WYY67 pKa = 10.32ASVANGASLSGTAFRR82 pKa = 11.84PNPYY86 pKa = 9.24PGYY89 pKa = 10.07RR90 pKa = 11.84SLFGDD95 pKa = 3.59TGLEE99 pKa = 3.81RR100 pKa = 11.84ANNVTSFRR108 pKa = 11.84YY109 pKa = 10.03ASTCVGLYY117 pKa = 6.81PTSNMQQFAGSISVWKK133 pKa = 10.35FPMEE137 pKa = 5.3LINQIVGVGTPVVGQPCWVLNGTDD161 pKa = 3.3NFSGIGYY168 pKa = 9.76EE169 pKa = 4.02NYY171 pKa = 9.3TDD173 pKa = 4.07SFIKK177 pKa = 10.54GMYY180 pKa = 8.48SQSVCNEE187 pKa = 3.51PDD189 pKa = 3.15FEE191 pKa = 4.13FRR193 pKa = 11.84PIIEE197 pKa = 4.88EE198 pKa = 3.59IGAIPSGVITATQAGMFCTFATSQPAAGQLCGIVGMGTMDD238 pKa = 5.36CILIKK243 pKa = 10.7VATPVGATNSAVLKK257 pKa = 8.96TWACVEE263 pKa = 3.93YY264 pKa = 10.66RR265 pKa = 11.84PNTNSALYY273 pKa = 10.06SYY275 pKa = 10.91AHH277 pKa = 7.35DD278 pKa = 4.3SPAPDD283 pKa = 4.07LMALAAYY290 pKa = 10.19RR291 pKa = 11.84EE292 pKa = 4.13IAKK295 pKa = 9.59NVPIAVPCSQNEE307 pKa = 4.43GFWKK311 pKa = 10.25RR312 pKa = 11.84VLTILRR318 pKa = 11.84GGLSLTSKK326 pKa = 10.48IPGPIGMTATGVSGIMDD343 pKa = 4.46MMTGLATT350 pKa = 3.64
Molecular weight: 37.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGU5|A0A1L3KGU5_9VIRU Capsid protein beta OS=Hubei unio douglasiae virus 1 OX=1923321 PE=4 SV=1
MM1 pKa = 7.65DD2 pKa = 4.36QFIEE6 pKa = 4.56CYY8 pKa = 9.74YY9 pKa = 10.35IEE11 pKa = 6.13LIFCTIILYY20 pKa = 10.23LVLVLQRR27 pKa = 11.84VKK29 pKa = 10.92EE30 pKa = 4.23KK31 pKa = 10.65ILASHH36 pKa = 6.49WNKK39 pKa = 10.15KK40 pKa = 7.17KK41 pKa = 10.43QRR43 pKa = 11.84ALTAQHH49 pKa = 6.3ISTRR53 pKa = 11.84VVRR56 pKa = 11.84YY57 pKa = 9.46ASATLRR63 pKa = 11.84TRR65 pKa = 11.84FQRR68 pKa = 11.84QMEE71 pKa = 4.32RR72 pKa = 11.84VHH74 pKa = 7.31SSVRR78 pKa = 11.84VGHH81 pKa = 5.49SHH83 pKa = 6.15QSAASEE89 pKa = 4.14RR90 pKa = 11.84NSATEE95 pKa = 3.5TMLAVVRR102 pKa = 11.84KK103 pKa = 9.77LGMIPYY109 pKa = 9.33VISPSPRR116 pKa = 11.84EE117 pKa = 3.85KK118 pKa = 10.92DD119 pKa = 3.07CDD121 pKa = 4.29GIRR124 pKa = 11.84DD125 pKa = 4.21YY126 pKa = 10.85YY127 pKa = 10.51TLADD131 pKa = 4.59LRR133 pKa = 11.84CQTKK137 pKa = 10.45NDD139 pKa = 5.17PITDD143 pKa = 3.27NHH145 pKa = 6.88VIIMTDD151 pKa = 2.23VDD153 pKa = 4.41YY154 pKa = 11.05YY155 pKa = 11.87VNMDD159 pKa = 4.48EE160 pKa = 4.65ILSHH164 pKa = 6.05GRR166 pKa = 11.84PVLLYY171 pKa = 8.36TFQPTKK177 pKa = 10.51VAGPVKK183 pKa = 10.41NGYY186 pKa = 7.72FTIEE190 pKa = 4.31HH191 pKa = 6.47DD192 pKa = 3.57TVTYY196 pKa = 9.76HH197 pKa = 6.38VKK199 pKa = 10.18GGKK202 pKa = 8.57DD203 pKa = 3.28VKK205 pKa = 10.8HH206 pKa = 6.48KK207 pKa = 9.64IWNYY211 pKa = 10.44SVDD214 pKa = 3.76TIYY217 pKa = 10.87TPNIVSGWFEE227 pKa = 4.4TYY229 pKa = 10.16FYY231 pKa = 10.85AITDD235 pKa = 3.79TLGEE239 pKa = 4.27LTPIDD244 pKa = 4.07GFFGLLSGLGKK255 pKa = 8.93LTPFHH260 pKa = 7.19KK261 pKa = 10.48EE262 pKa = 3.44INNFLYY268 pKa = 9.19QTVTVSTIDD277 pKa = 3.12QFYY280 pKa = 10.39LSEE283 pKa = 4.45HH284 pKa = 5.66RR285 pKa = 11.84TITTIVPYY293 pKa = 10.23ARR295 pKa = 11.84CPTDD299 pKa = 4.6IIDD302 pKa = 4.05TLAPATRR309 pKa = 11.84LVRR312 pKa = 11.84MNYY315 pKa = 8.98QVPDD319 pKa = 3.66EE320 pKa = 4.35DD321 pKa = 5.45HH322 pKa = 7.21IEE324 pKa = 4.28PKK326 pKa = 10.34NDD328 pKa = 2.91LRR330 pKa = 11.84FNCLVHH336 pKa = 8.51ISDD339 pKa = 4.11SGPNISVGIAGEE351 pKa = 4.18LAQATLPLSLFEE363 pKa = 4.58SLRR366 pKa = 11.84TAHH369 pKa = 6.63SLAKK373 pKa = 10.31ARR375 pKa = 11.84ALSDD379 pKa = 3.18TVRR382 pKa = 11.84RR383 pKa = 11.84SKK385 pKa = 11.21LDD387 pKa = 3.56NNEE390 pKa = 4.03SATLHH395 pKa = 6.2CFLTGVQKK403 pKa = 9.37STPDD407 pKa = 3.19IVHH410 pKa = 6.14SPGLFSRR417 pKa = 11.84HH418 pKa = 5.42FNSVDD423 pKa = 3.49QQDD426 pKa = 3.53DD427 pKa = 4.29TIPYY431 pKa = 10.57DD432 pKa = 3.33NGKK435 pKa = 10.39EE436 pKa = 4.05YY437 pKa = 10.99ARR439 pKa = 11.84DD440 pKa = 3.68YY441 pKa = 11.65APGPLSDD448 pKa = 3.36TAVFPTEE455 pKa = 4.07SSNNDD460 pKa = 2.85KK461 pKa = 10.33ATIRR465 pKa = 11.84GRR467 pKa = 11.84IDD469 pKa = 2.87IPQRR473 pKa = 11.84IANGTQYY480 pKa = 10.81IKK482 pKa = 10.73PQFLKK487 pKa = 10.33FAHH490 pKa = 6.29EE491 pKa = 4.3FVSIILKK498 pKa = 8.57NTTEE502 pKa = 4.4RR503 pKa = 11.84LGQPYY508 pKa = 10.69SNDD511 pKa = 3.24YY512 pKa = 11.02VIEE515 pKa = 4.21KK516 pKa = 9.87QNKK519 pKa = 6.96PRR521 pKa = 11.84QRR523 pKa = 11.84QRR525 pKa = 11.84NDD527 pKa = 2.57QGLFHH532 pKa = 7.38VIDD535 pKa = 3.92KK536 pKa = 10.1FVVRR540 pKa = 11.84AFQKK544 pKa = 10.26RR545 pKa = 11.84EE546 pKa = 3.82AYY548 pKa = 10.23NGPNYY553 pKa = 9.21PRR555 pKa = 11.84NISTVPGYY563 pKa = 10.99HH564 pKa = 5.35NTRR567 pKa = 11.84LSGFTYY573 pKa = 10.42AFKK576 pKa = 10.82DD577 pKa = 3.44AVLRR581 pKa = 11.84THH583 pKa = 7.41DD584 pKa = 3.54WYY586 pKa = 11.1QPCRR590 pKa = 11.84TPEE593 pKa = 3.97EE594 pKa = 3.75IAANVHH600 pKa = 6.37ALAQKK605 pKa = 10.47HH606 pKa = 5.46EE607 pKa = 4.3YY608 pKa = 10.19LVEE611 pKa = 3.87TDD613 pKa = 3.42YY614 pKa = 11.89SKK616 pKa = 11.44FDD618 pKa = 3.42GTVTRR623 pKa = 11.84WIRR626 pKa = 11.84EE627 pKa = 3.97NVEE630 pKa = 4.24FPCYY634 pKa = 9.93LRR636 pKa = 11.84WVAPEE641 pKa = 4.57HH642 pKa = 6.81RR643 pKa = 11.84DD644 pKa = 3.58EE645 pKa = 5.99LNTLLLSEE653 pKa = 5.12LDD655 pKa = 4.16APGTTNTISYY665 pKa = 10.75SPGCSRR671 pKa = 11.84LSGSPLTTDD680 pKa = 4.26GNTICNAFVSYY691 pKa = 10.51CAARR695 pKa = 11.84SMKK698 pKa = 9.13YY699 pKa = 8.7TEE701 pKa = 5.2AEE703 pKa = 3.97AFSDD707 pKa = 4.13IGLCYY712 pKa = 10.5GDD714 pKa = 4.55DD715 pKa = 3.81GLRR718 pKa = 11.84GGNVANTKK726 pKa = 9.33LRR728 pKa = 11.84KK729 pKa = 8.5IASRR733 pKa = 11.84LGFDD737 pKa = 4.45LRR739 pKa = 11.84VCNRR743 pKa = 11.84AKK745 pKa = 10.61AGSPVSFLSRR755 pKa = 11.84VFADD759 pKa = 3.75PWTSPASIQSPLRR772 pKa = 11.84TLLKK776 pKa = 10.05IHH778 pKa = 5.65TTVDD782 pKa = 2.95RR783 pKa = 11.84RR784 pKa = 11.84TKK786 pKa = 10.64LSVVGAAKK794 pKa = 7.23TTAYY798 pKa = 10.67LVTDD802 pKa = 3.28SRR804 pKa = 11.84TPFISDD810 pKa = 3.01WCRR813 pKa = 11.84AYY815 pKa = 10.7QKK817 pKa = 10.45AAKK820 pKa = 8.52AQPVANFNDD829 pKa = 3.94LDD831 pKa = 5.33DD832 pKa = 4.1IPFWVLDD839 pKa = 3.99TDD841 pKa = 4.27NLAHH845 pKa = 7.34PWPQDD850 pKa = 3.36DD851 pKa = 4.55PEE853 pKa = 5.82NYY855 pKa = 8.66MDD857 pKa = 4.99IVATEE862 pKa = 4.16LGITASEE869 pKa = 4.08LSDD872 pKa = 4.21HH873 pKa = 6.95IKK875 pKa = 10.75LLNEE879 pKa = 3.86YY880 pKa = 9.89SGDD883 pKa = 3.77VAGLPVLQTTIDD895 pKa = 3.72QTPKK899 pKa = 10.42LAVVLDD905 pKa = 4.17GEE907 pKa = 4.48IHH909 pKa = 7.21AGPNLQDD916 pKa = 4.27DD917 pKa = 4.54SNKK920 pKa = 10.46HH921 pKa = 5.05GIARR925 pKa = 11.84QDD927 pKa = 3.29VPVAQHH933 pKa = 5.95RR934 pKa = 11.84RR935 pKa = 11.84SGAVEE940 pKa = 3.8RR941 pKa = 11.84EE942 pKa = 3.66NRR944 pKa = 11.84ARR946 pKa = 11.84PPHH949 pKa = 6.49VSTAPRR955 pKa = 11.84RR956 pKa = 11.84GPQSNYY962 pKa = 9.43PRR964 pKa = 11.84GSTNPHH970 pKa = 6.51KK971 pKa = 10.44DD972 pKa = 3.11SKK974 pKa = 11.13EE975 pKa = 3.82RR976 pKa = 11.84TDD978 pKa = 3.71PVSSKK983 pKa = 10.58HH984 pKa = 6.08RR985 pKa = 11.84GADD988 pKa = 3.04QTHH991 pKa = 6.61PPKK994 pKa = 10.86CEE996 pKa = 3.73ANGRR1000 pKa = 11.84RR1001 pKa = 11.84SGKK1004 pKa = 7.94TRR1006 pKa = 11.84GRR1008 pKa = 11.84TGKK1011 pKa = 10.61SNAATDD1017 pKa = 3.78QSNHH1021 pKa = 4.71GSAAGGRR1028 pKa = 11.84QSKK1031 pKa = 7.98ITQGLAKK1038 pKa = 9.75TDD1040 pKa = 3.31
MM1 pKa = 7.65DD2 pKa = 4.36QFIEE6 pKa = 4.56CYY8 pKa = 9.74YY9 pKa = 10.35IEE11 pKa = 6.13LIFCTIILYY20 pKa = 10.23LVLVLQRR27 pKa = 11.84VKK29 pKa = 10.92EE30 pKa = 4.23KK31 pKa = 10.65ILASHH36 pKa = 6.49WNKK39 pKa = 10.15KK40 pKa = 7.17KK41 pKa = 10.43QRR43 pKa = 11.84ALTAQHH49 pKa = 6.3ISTRR53 pKa = 11.84VVRR56 pKa = 11.84YY57 pKa = 9.46ASATLRR63 pKa = 11.84TRR65 pKa = 11.84FQRR68 pKa = 11.84QMEE71 pKa = 4.32RR72 pKa = 11.84VHH74 pKa = 7.31SSVRR78 pKa = 11.84VGHH81 pKa = 5.49SHH83 pKa = 6.15QSAASEE89 pKa = 4.14RR90 pKa = 11.84NSATEE95 pKa = 3.5TMLAVVRR102 pKa = 11.84KK103 pKa = 9.77LGMIPYY109 pKa = 9.33VISPSPRR116 pKa = 11.84EE117 pKa = 3.85KK118 pKa = 10.92DD119 pKa = 3.07CDD121 pKa = 4.29GIRR124 pKa = 11.84DD125 pKa = 4.21YY126 pKa = 10.85YY127 pKa = 10.51TLADD131 pKa = 4.59LRR133 pKa = 11.84CQTKK137 pKa = 10.45NDD139 pKa = 5.17PITDD143 pKa = 3.27NHH145 pKa = 6.88VIIMTDD151 pKa = 2.23VDD153 pKa = 4.41YY154 pKa = 11.05YY155 pKa = 11.87VNMDD159 pKa = 4.48EE160 pKa = 4.65ILSHH164 pKa = 6.05GRR166 pKa = 11.84PVLLYY171 pKa = 8.36TFQPTKK177 pKa = 10.51VAGPVKK183 pKa = 10.41NGYY186 pKa = 7.72FTIEE190 pKa = 4.31HH191 pKa = 6.47DD192 pKa = 3.57TVTYY196 pKa = 9.76HH197 pKa = 6.38VKK199 pKa = 10.18GGKK202 pKa = 8.57DD203 pKa = 3.28VKK205 pKa = 10.8HH206 pKa = 6.48KK207 pKa = 9.64IWNYY211 pKa = 10.44SVDD214 pKa = 3.76TIYY217 pKa = 10.87TPNIVSGWFEE227 pKa = 4.4TYY229 pKa = 10.16FYY231 pKa = 10.85AITDD235 pKa = 3.79TLGEE239 pKa = 4.27LTPIDD244 pKa = 4.07GFFGLLSGLGKK255 pKa = 8.93LTPFHH260 pKa = 7.19KK261 pKa = 10.48EE262 pKa = 3.44INNFLYY268 pKa = 9.19QTVTVSTIDD277 pKa = 3.12QFYY280 pKa = 10.39LSEE283 pKa = 4.45HH284 pKa = 5.66RR285 pKa = 11.84TITTIVPYY293 pKa = 10.23ARR295 pKa = 11.84CPTDD299 pKa = 4.6IIDD302 pKa = 4.05TLAPATRR309 pKa = 11.84LVRR312 pKa = 11.84MNYY315 pKa = 8.98QVPDD319 pKa = 3.66EE320 pKa = 4.35DD321 pKa = 5.45HH322 pKa = 7.21IEE324 pKa = 4.28PKK326 pKa = 10.34NDD328 pKa = 2.91LRR330 pKa = 11.84FNCLVHH336 pKa = 8.51ISDD339 pKa = 4.11SGPNISVGIAGEE351 pKa = 4.18LAQATLPLSLFEE363 pKa = 4.58SLRR366 pKa = 11.84TAHH369 pKa = 6.63SLAKK373 pKa = 10.31ARR375 pKa = 11.84ALSDD379 pKa = 3.18TVRR382 pKa = 11.84RR383 pKa = 11.84SKK385 pKa = 11.21LDD387 pKa = 3.56NNEE390 pKa = 4.03SATLHH395 pKa = 6.2CFLTGVQKK403 pKa = 9.37STPDD407 pKa = 3.19IVHH410 pKa = 6.14SPGLFSRR417 pKa = 11.84HH418 pKa = 5.42FNSVDD423 pKa = 3.49QQDD426 pKa = 3.53DD427 pKa = 4.29TIPYY431 pKa = 10.57DD432 pKa = 3.33NGKK435 pKa = 10.39EE436 pKa = 4.05YY437 pKa = 10.99ARR439 pKa = 11.84DD440 pKa = 3.68YY441 pKa = 11.65APGPLSDD448 pKa = 3.36TAVFPTEE455 pKa = 4.07SSNNDD460 pKa = 2.85KK461 pKa = 10.33ATIRR465 pKa = 11.84GRR467 pKa = 11.84IDD469 pKa = 2.87IPQRR473 pKa = 11.84IANGTQYY480 pKa = 10.81IKK482 pKa = 10.73PQFLKK487 pKa = 10.33FAHH490 pKa = 6.29EE491 pKa = 4.3FVSIILKK498 pKa = 8.57NTTEE502 pKa = 4.4RR503 pKa = 11.84LGQPYY508 pKa = 10.69SNDD511 pKa = 3.24YY512 pKa = 11.02VIEE515 pKa = 4.21KK516 pKa = 9.87QNKK519 pKa = 6.96PRR521 pKa = 11.84QRR523 pKa = 11.84QRR525 pKa = 11.84NDD527 pKa = 2.57QGLFHH532 pKa = 7.38VIDD535 pKa = 3.92KK536 pKa = 10.1FVVRR540 pKa = 11.84AFQKK544 pKa = 10.26RR545 pKa = 11.84EE546 pKa = 3.82AYY548 pKa = 10.23NGPNYY553 pKa = 9.21PRR555 pKa = 11.84NISTVPGYY563 pKa = 10.99HH564 pKa = 5.35NTRR567 pKa = 11.84LSGFTYY573 pKa = 10.42AFKK576 pKa = 10.82DD577 pKa = 3.44AVLRR581 pKa = 11.84THH583 pKa = 7.41DD584 pKa = 3.54WYY586 pKa = 11.1QPCRR590 pKa = 11.84TPEE593 pKa = 3.97EE594 pKa = 3.75IAANVHH600 pKa = 6.37ALAQKK605 pKa = 10.47HH606 pKa = 5.46EE607 pKa = 4.3YY608 pKa = 10.19LVEE611 pKa = 3.87TDD613 pKa = 3.42YY614 pKa = 11.89SKK616 pKa = 11.44FDD618 pKa = 3.42GTVTRR623 pKa = 11.84WIRR626 pKa = 11.84EE627 pKa = 3.97NVEE630 pKa = 4.24FPCYY634 pKa = 9.93LRR636 pKa = 11.84WVAPEE641 pKa = 4.57HH642 pKa = 6.81RR643 pKa = 11.84DD644 pKa = 3.58EE645 pKa = 5.99LNTLLLSEE653 pKa = 5.12LDD655 pKa = 4.16APGTTNTISYY665 pKa = 10.75SPGCSRR671 pKa = 11.84LSGSPLTTDD680 pKa = 4.26GNTICNAFVSYY691 pKa = 10.51CAARR695 pKa = 11.84SMKK698 pKa = 9.13YY699 pKa = 8.7TEE701 pKa = 5.2AEE703 pKa = 3.97AFSDD707 pKa = 4.13IGLCYY712 pKa = 10.5GDD714 pKa = 4.55DD715 pKa = 3.81GLRR718 pKa = 11.84GGNVANTKK726 pKa = 9.33LRR728 pKa = 11.84KK729 pKa = 8.5IASRR733 pKa = 11.84LGFDD737 pKa = 4.45LRR739 pKa = 11.84VCNRR743 pKa = 11.84AKK745 pKa = 10.61AGSPVSFLSRR755 pKa = 11.84VFADD759 pKa = 3.75PWTSPASIQSPLRR772 pKa = 11.84TLLKK776 pKa = 10.05IHH778 pKa = 5.65TTVDD782 pKa = 2.95RR783 pKa = 11.84RR784 pKa = 11.84TKK786 pKa = 10.64LSVVGAAKK794 pKa = 7.23TTAYY798 pKa = 10.67LVTDD802 pKa = 3.28SRR804 pKa = 11.84TPFISDD810 pKa = 3.01WCRR813 pKa = 11.84AYY815 pKa = 10.7QKK817 pKa = 10.45AAKK820 pKa = 8.52AQPVANFNDD829 pKa = 3.94LDD831 pKa = 5.33DD832 pKa = 4.1IPFWVLDD839 pKa = 3.99TDD841 pKa = 4.27NLAHH845 pKa = 7.34PWPQDD850 pKa = 3.36DD851 pKa = 4.55PEE853 pKa = 5.82NYY855 pKa = 8.66MDD857 pKa = 4.99IVATEE862 pKa = 4.16LGITASEE869 pKa = 4.08LSDD872 pKa = 4.21HH873 pKa = 6.95IKK875 pKa = 10.75LLNEE879 pKa = 3.86YY880 pKa = 9.89SGDD883 pKa = 3.77VAGLPVLQTTIDD895 pKa = 3.72QTPKK899 pKa = 10.42LAVVLDD905 pKa = 4.17GEE907 pKa = 4.48IHH909 pKa = 7.21AGPNLQDD916 pKa = 4.27DD917 pKa = 4.54SNKK920 pKa = 10.46HH921 pKa = 5.05GIARR925 pKa = 11.84QDD927 pKa = 3.29VPVAQHH933 pKa = 5.95RR934 pKa = 11.84RR935 pKa = 11.84SGAVEE940 pKa = 3.8RR941 pKa = 11.84EE942 pKa = 3.66NRR944 pKa = 11.84ARR946 pKa = 11.84PPHH949 pKa = 6.49VSTAPRR955 pKa = 11.84RR956 pKa = 11.84GPQSNYY962 pKa = 9.43PRR964 pKa = 11.84GSTNPHH970 pKa = 6.51KK971 pKa = 10.44DD972 pKa = 3.11SKK974 pKa = 11.13EE975 pKa = 3.82RR976 pKa = 11.84TDD978 pKa = 3.71PVSSKK983 pKa = 10.58HH984 pKa = 6.08RR985 pKa = 11.84GADD988 pKa = 3.04QTHH991 pKa = 6.61PPKK994 pKa = 10.86CEE996 pKa = 3.73ANGRR1000 pKa = 11.84RR1001 pKa = 11.84SGKK1004 pKa = 7.94TRR1006 pKa = 11.84GRR1008 pKa = 11.84TGKK1011 pKa = 10.61SNAATDD1017 pKa = 3.78QSNHH1021 pKa = 4.71GSAAGGRR1028 pKa = 11.84QSKK1031 pKa = 7.98ITQGLAKK1038 pKa = 9.75TDD1040 pKa = 3.31
Molecular weight: 116.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1544 |
154 |
1040 |
514.7 |
57.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.578 ± 0.697 | 1.619 ± 0.397 |
5.894 ± 1.041 | 4.275 ± 0.615 |
3.368 ± 0.711 | 6.412 ± 1.633 |
2.591 ± 0.989 | 5.829 ± 0.183 |
4.534 ± 0.613 | 8.355 ± 1.918 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.813 ± 1.081 | 4.793 ± 0.124 |
5.829 ± 0.453 | 4.016 ± 0.522 |
5.959 ± 1.129 | 7.124 ± 0.214 |
8.225 ± 0.081 | 6.736 ± 0.418 |
1.101 ± 0.156 | 3.951 ± 0.422 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
