
bacterium HR41
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
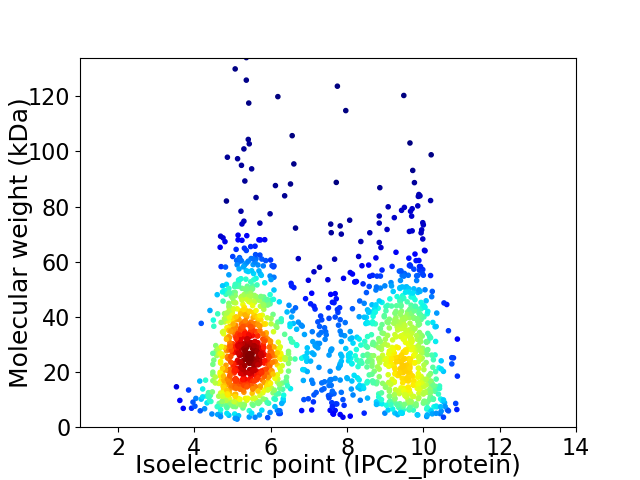
Virtual 2D-PAGE plot for 1586 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H6B7P1|A0A2H6B7P1_9BACT Uncharacterized protein OS=bacterium HR41 OX=2035436 GN=HRbin41_01029 PE=4 SV=1
MM1 pKa = 7.69EE2 pKa = 4.28SHH4 pKa = 6.46EE5 pKa = 4.36LEE7 pKa = 4.38KK8 pKa = 10.17THH10 pKa = 7.03ADD12 pKa = 3.51SGGSAGDD19 pKa = 3.65PSAAAGDD26 pKa = 3.98GPPTVEE32 pKa = 4.35EE33 pKa = 3.99VRR35 pKa = 11.84EE36 pKa = 4.0ALTNVIDD43 pKa = 4.45PEE45 pKa = 5.32LGLDD49 pKa = 3.98FVEE52 pKa = 5.65LGLIYY57 pKa = 9.95DD58 pKa = 3.99IQVEE62 pKa = 4.2DD63 pKa = 3.85GDD65 pKa = 4.51VYY67 pKa = 10.49VTYY70 pKa = 10.2TLTSPGCPIGPHH82 pKa = 6.17IADD85 pKa = 3.55QIEE88 pKa = 4.5EE89 pKa = 4.41FVSEE93 pKa = 4.2LPGVQRR99 pKa = 11.84VYY101 pKa = 10.99PKK103 pKa = 10.11LTFDD107 pKa = 4.64PPWTPDD113 pKa = 3.22MMSEE117 pKa = 4.08DD118 pKa = 3.6AKK120 pKa = 10.95FALGYY125 pKa = 10.59
MM1 pKa = 7.69EE2 pKa = 4.28SHH4 pKa = 6.46EE5 pKa = 4.36LEE7 pKa = 4.38KK8 pKa = 10.17THH10 pKa = 7.03ADD12 pKa = 3.51SGGSAGDD19 pKa = 3.65PSAAAGDD26 pKa = 3.98GPPTVEE32 pKa = 4.35EE33 pKa = 3.99VRR35 pKa = 11.84EE36 pKa = 4.0ALTNVIDD43 pKa = 4.45PEE45 pKa = 5.32LGLDD49 pKa = 3.98FVEE52 pKa = 5.65LGLIYY57 pKa = 9.95DD58 pKa = 3.99IQVEE62 pKa = 4.2DD63 pKa = 3.85GDD65 pKa = 4.51VYY67 pKa = 10.49VTYY70 pKa = 10.2TLTSPGCPIGPHH82 pKa = 6.17IADD85 pKa = 3.55QIEE88 pKa = 4.5EE89 pKa = 4.41FVSEE93 pKa = 4.2LPGVQRR99 pKa = 11.84VYY101 pKa = 10.99PKK103 pKa = 10.11LTFDD107 pKa = 4.64PPWTPDD113 pKa = 3.22MMSEE117 pKa = 4.08DD118 pKa = 3.6AKK120 pKa = 10.95FALGYY125 pKa = 10.59
Molecular weight: 13.48 kDa
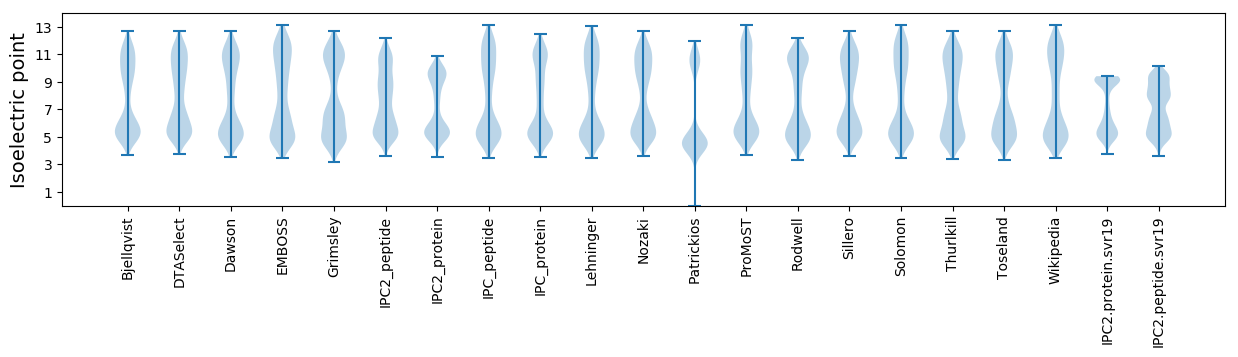
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H6B883|A0A2H6B883_9BACT UPF0109 protein HRbin41_01221 OS=bacterium HR41 OX=2035436 GN=HRbin41_01221 PE=3 SV=1
MM1 pKa = 7.49GAPRR5 pKa = 11.84PPRR8 pKa = 11.84ATKK11 pKa = 10.57VPATTSRR18 pKa = 11.84APRR21 pKa = 11.84ATPQLAAALPAPRR34 pKa = 11.84AAKK37 pKa = 10.36LGATARR43 pKa = 11.84RR44 pKa = 11.84STSSCRR50 pKa = 11.84RR51 pKa = 11.84WASRR55 pKa = 11.84CKK57 pKa = 10.0RR58 pKa = 11.84APSSSGSSASATRR71 pKa = 11.84SARR74 pKa = 11.84DD75 pKa = 3.32SRR77 pKa = 11.84SSRR80 pKa = 11.84FRR82 pKa = 11.84PIRR85 pKa = 11.84STPRR89 pKa = 11.84SPHH92 pKa = 6.69PPTEE96 pKa = 4.05PSPRR100 pKa = 11.84SSSRR104 pKa = 11.84ATRR107 pKa = 11.84RR108 pKa = 11.84SPSVPSSAGSQRR120 pKa = 11.84VRR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84RR126 pKa = 11.84SSRR129 pKa = 11.84ATLAVKK135 pKa = 9.83DD136 pKa = 3.7APRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84SRR143 pKa = 11.84VLRR146 pKa = 11.84VGRR149 pKa = 11.84RR150 pKa = 11.84PGPRR154 pKa = 11.84LSRR157 pKa = 11.84RR158 pKa = 11.84AAPGSATGLQRR169 pKa = 11.84RR170 pKa = 11.84PSLAVRR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84LTVSTSRR185 pKa = 11.84ACRR188 pKa = 11.84AAGPGVASPKK198 pKa = 9.32RR199 pKa = 11.84TSCVQQAARR208 pKa = 11.84APPAAMAQQPTALATPAAPAWLPLARR234 pKa = 11.84RR235 pKa = 11.84ALRR238 pKa = 11.84RR239 pKa = 11.84HH240 pKa = 5.91RR241 pKa = 11.84PFGRR245 pKa = 11.84CRR247 pKa = 11.84CADD250 pKa = 3.42RR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84WPATWSAAARR264 pKa = 11.84SRR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84ASARR273 pKa = 11.84SRR275 pKa = 11.84STSSTGAGASSRR287 pKa = 11.84PPANVCRR294 pKa = 11.84SLTT297 pKa = 3.51
MM1 pKa = 7.49GAPRR5 pKa = 11.84PPRR8 pKa = 11.84ATKK11 pKa = 10.57VPATTSRR18 pKa = 11.84APRR21 pKa = 11.84ATPQLAAALPAPRR34 pKa = 11.84AAKK37 pKa = 10.36LGATARR43 pKa = 11.84RR44 pKa = 11.84STSSCRR50 pKa = 11.84RR51 pKa = 11.84WASRR55 pKa = 11.84CKK57 pKa = 10.0RR58 pKa = 11.84APSSSGSSASATRR71 pKa = 11.84SARR74 pKa = 11.84DD75 pKa = 3.32SRR77 pKa = 11.84SSRR80 pKa = 11.84FRR82 pKa = 11.84PIRR85 pKa = 11.84STPRR89 pKa = 11.84SPHH92 pKa = 6.69PPTEE96 pKa = 4.05PSPRR100 pKa = 11.84SSSRR104 pKa = 11.84ATRR107 pKa = 11.84RR108 pKa = 11.84SPSVPSSAGSQRR120 pKa = 11.84VRR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84RR126 pKa = 11.84SSRR129 pKa = 11.84ATLAVKK135 pKa = 9.83DD136 pKa = 3.7APRR139 pKa = 11.84RR140 pKa = 11.84RR141 pKa = 11.84SRR143 pKa = 11.84VLRR146 pKa = 11.84VGRR149 pKa = 11.84RR150 pKa = 11.84PGPRR154 pKa = 11.84LSRR157 pKa = 11.84RR158 pKa = 11.84AAPGSATGLQRR169 pKa = 11.84RR170 pKa = 11.84PSLAVRR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84LTVSTSRR185 pKa = 11.84ACRR188 pKa = 11.84AAGPGVASPKK198 pKa = 9.32RR199 pKa = 11.84TSCVQQAARR208 pKa = 11.84APPAAMAQQPTALATPAAPAWLPLARR234 pKa = 11.84RR235 pKa = 11.84ALRR238 pKa = 11.84RR239 pKa = 11.84HH240 pKa = 5.91RR241 pKa = 11.84PFGRR245 pKa = 11.84CRR247 pKa = 11.84CADD250 pKa = 3.42RR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84WPATWSAAARR264 pKa = 11.84SRR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84ASARR273 pKa = 11.84SRR275 pKa = 11.84STSSTGAGASSRR287 pKa = 11.84PPANVCRR294 pKa = 11.84SLTT297 pKa = 3.51
Molecular weight: 31.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
457819 |
29 |
1209 |
288.7 |
31.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.74 ± 0.107 | 1.027 ± 0.019 |
5.284 ± 0.051 | 6.742 ± 0.078 |
2.88 ± 0.034 | 8.816 ± 0.059 |
1.97 ± 0.032 | 3.38 ± 0.045 |
1.661 ± 0.037 | 10.633 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.347 ± 0.024 | 1.537 ± 0.025 |
5.763 ± 0.044 | 2.55 ± 0.033 |
10.832 ± 0.074 | 4.61 ± 0.042 |
4.528 ± 0.037 | 8.758 ± 0.058 |
1.176 ± 0.026 | 1.768 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
