
Aquisalibacillus elongatus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Aquisalibacillus
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
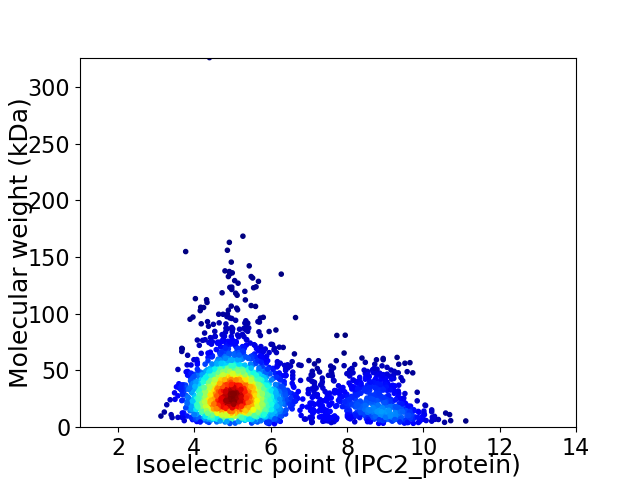
Virtual 2D-PAGE plot for 2958 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
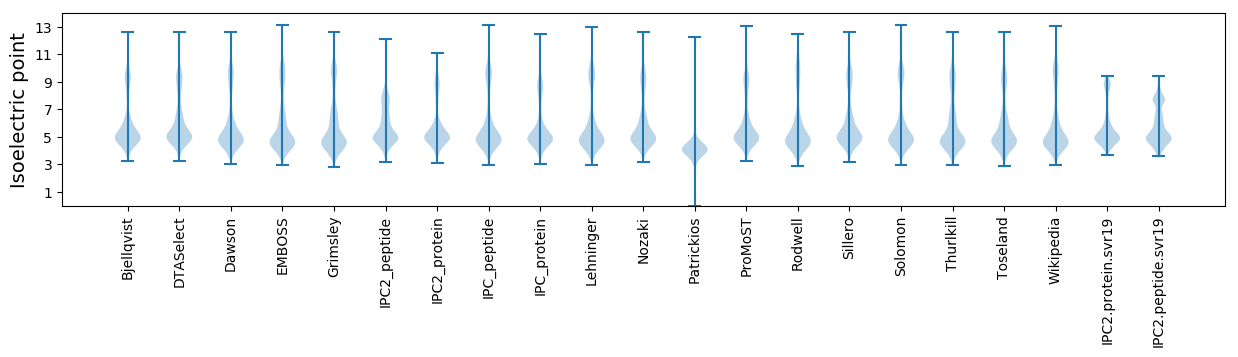
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N5CDP6|A0A3N5CDP6_9BACI Aldehyde dehydrogenase (Acceptor) OS=Aquisalibacillus elongatus OX=485577 GN=EDC24_0122 PE=3 SV=1
MM1 pKa = 7.39SKK3 pKa = 10.68SLFGKK8 pKa = 10.08FLFVLMLAFVLLIAACNSDD27 pKa = 3.3EE28 pKa = 4.29GGEE31 pKa = 4.08DD32 pKa = 3.9SSDD35 pKa = 3.43NDD37 pKa = 5.12GDD39 pKa = 3.71DD40 pKa = 3.86TEE42 pKa = 5.04QNEE45 pKa = 4.7SDD47 pKa = 5.35DD48 pKa = 5.04DD49 pKa = 5.46DD50 pKa = 6.08SDD52 pKa = 4.29SDD54 pKa = 3.93SDD56 pKa = 4.3SDD58 pKa = 4.08QEE60 pKa = 4.92LYY62 pKa = 11.06SIDD65 pKa = 4.54DD66 pKa = 3.82FEE68 pKa = 6.46SYY70 pKa = 9.79TGEE73 pKa = 4.0GDD75 pKa = 4.32AIDD78 pKa = 4.54GGSLTFGLVSQSPFEE93 pKa = 4.34GTLNWQFYY101 pKa = 10.42SGNPDD106 pKa = 3.39SQVLSWFDD114 pKa = 3.67EE115 pKa = 4.15SLLQYY120 pKa = 10.79DD121 pKa = 3.66EE122 pKa = 4.93NYY124 pKa = 10.97NVTNEE129 pKa = 3.82GAASYY134 pKa = 9.66EE135 pKa = 4.19VNEE138 pKa = 4.47EE139 pKa = 4.01DD140 pKa = 4.53SSITFTIEE148 pKa = 5.8DD149 pKa = 3.7DD150 pKa = 4.51VKK152 pKa = 9.47WHH154 pKa = 7.13DD155 pKa = 3.9GEE157 pKa = 4.63EE158 pKa = 4.22VTAEE162 pKa = 3.79DD163 pKa = 2.96WAYY166 pKa = 9.07SYY168 pKa = 10.94EE169 pKa = 4.56VIMHH173 pKa = 7.09PDD175 pKa = 3.17YY176 pKa = 11.6DD177 pKa = 4.06GVRR180 pKa = 11.84GSSAGFAYY188 pKa = 10.74LEE190 pKa = 4.47GAQEE194 pKa = 4.17YY195 pKa = 10.0KK196 pKa = 10.74AGEE199 pKa = 4.16ADD201 pKa = 3.94SISGIEE207 pKa = 3.97IEE209 pKa = 4.38GEE211 pKa = 3.77KK212 pKa = 10.23TITFNYY218 pKa = 9.54TEE220 pKa = 4.64LTPSLLTGGVWGYY233 pKa = 10.41PMAKK237 pKa = 9.57HH238 pKa = 5.58VFEE241 pKa = 6.08DD242 pKa = 3.92IPVEE246 pKa = 4.24DD247 pKa = 4.18MASSPEE253 pKa = 3.95VRR255 pKa = 11.84EE256 pKa = 3.99EE257 pKa = 4.33PIGFGPYY264 pKa = 9.31KK265 pKa = 10.41VEE267 pKa = 3.79NVVPGEE273 pKa = 4.07SVTYY277 pKa = 10.73SKK279 pKa = 11.3FEE281 pKa = 4.61DD282 pKa = 3.82YY283 pKa = 10.28WRR285 pKa = 11.84GEE287 pKa = 4.06PNLDD291 pKa = 3.25EE292 pKa = 4.37VTLKK296 pKa = 10.7VVSPSNVANALEE308 pKa = 4.4TGDD311 pKa = 3.83VDD313 pKa = 5.66LVDD316 pKa = 5.12SFPADD321 pKa = 3.16QVVDD325 pKa = 4.41NIDD328 pKa = 3.55MPNVQWLGMTDD339 pKa = 3.59LAYY342 pKa = 9.64TYY344 pKa = 10.46IGFKK348 pKa = 10.63LGDD351 pKa = 3.36WNADD355 pKa = 3.41EE356 pKa = 6.22GEE358 pKa = 4.27VNYY361 pKa = 10.89KK362 pKa = 9.83GDD364 pKa = 3.76EE365 pKa = 3.99MKK367 pKa = 10.54MGDD370 pKa = 3.45KK371 pKa = 10.46KK372 pKa = 10.76LRR374 pKa = 11.84KK375 pKa = 9.43AMWYY379 pKa = 10.12AVDD382 pKa = 3.76NEE384 pKa = 4.83TVGEE388 pKa = 4.03RR389 pKa = 11.84FYY391 pKa = 11.8NGLRR395 pKa = 11.84WNGTTLIPPSHH406 pKa = 6.93PAWHH410 pKa = 7.62DD411 pKa = 3.59DD412 pKa = 3.91SIEE415 pKa = 4.12TPTYY419 pKa = 11.03DD420 pKa = 3.32EE421 pKa = 5.35DD422 pKa = 4.35KK423 pKa = 11.33ANEE426 pKa = 4.09LLDD429 pKa = 4.01DD430 pKa = 5.02AGYY433 pKa = 11.18KK434 pKa = 8.79DD435 pKa = 4.26TNDD438 pKa = 3.23DD439 pKa = 4.23GYY441 pKa = 11.32RR442 pKa = 11.84EE443 pKa = 4.22TPDD446 pKa = 3.85GEE448 pKa = 3.97EE449 pKa = 3.83LVINFASMDD458 pKa = 3.74SSDD461 pKa = 4.53LSEE464 pKa = 4.54PLANYY469 pKa = 8.64YY470 pKa = 8.97IQAWDD475 pKa = 3.52KK476 pKa = 11.58VGLNVQKK483 pKa = 10.93LDD485 pKa = 3.75GRR487 pKa = 11.84LHH489 pKa = 6.38EE490 pKa = 5.13FNSFYY495 pKa = 11.28DD496 pKa = 3.53KK497 pKa = 11.09VGQTGEE503 pKa = 4.45DD504 pKa = 3.8DD505 pKa = 4.21PNVDD509 pKa = 3.14VFMGAWSVGYY519 pKa = 10.32DD520 pKa = 3.43VDD522 pKa = 4.11PSGLYY527 pKa = 10.68GRR529 pKa = 11.84DD530 pKa = 2.84ALFNFTRR537 pKa = 11.84WANEE541 pKa = 3.78EE542 pKa = 3.85NDD544 pKa = 3.51EE545 pKa = 4.4LLAEE549 pKa = 4.61GLSEE553 pKa = 3.92EE554 pKa = 5.02AFDD557 pKa = 3.5TEE559 pKa = 4.21YY560 pKa = 11.1RR561 pKa = 11.84QDD563 pKa = 5.0VYY565 pKa = 11.22SQWQEE570 pKa = 3.7LMVEE574 pKa = 4.39EE575 pKa = 4.74VPVFPTLYY583 pKa = 9.46RR584 pKa = 11.84AMMVPANEE592 pKa = 3.76RR593 pKa = 11.84VVNYY597 pKa = 9.63DD598 pKa = 2.76ISVGSGVYY606 pKa = 10.09LYY608 pKa = 10.62EE609 pKa = 5.06LGVTQEE615 pKa = 4.21EE616 pKa = 4.86PVVEE620 pKa = 4.24
MM1 pKa = 7.39SKK3 pKa = 10.68SLFGKK8 pKa = 10.08FLFVLMLAFVLLIAACNSDD27 pKa = 3.3EE28 pKa = 4.29GGEE31 pKa = 4.08DD32 pKa = 3.9SSDD35 pKa = 3.43NDD37 pKa = 5.12GDD39 pKa = 3.71DD40 pKa = 3.86TEE42 pKa = 5.04QNEE45 pKa = 4.7SDD47 pKa = 5.35DD48 pKa = 5.04DD49 pKa = 5.46DD50 pKa = 6.08SDD52 pKa = 4.29SDD54 pKa = 3.93SDD56 pKa = 4.3SDD58 pKa = 4.08QEE60 pKa = 4.92LYY62 pKa = 11.06SIDD65 pKa = 4.54DD66 pKa = 3.82FEE68 pKa = 6.46SYY70 pKa = 9.79TGEE73 pKa = 4.0GDD75 pKa = 4.32AIDD78 pKa = 4.54GGSLTFGLVSQSPFEE93 pKa = 4.34GTLNWQFYY101 pKa = 10.42SGNPDD106 pKa = 3.39SQVLSWFDD114 pKa = 3.67EE115 pKa = 4.15SLLQYY120 pKa = 10.79DD121 pKa = 3.66EE122 pKa = 4.93NYY124 pKa = 10.97NVTNEE129 pKa = 3.82GAASYY134 pKa = 9.66EE135 pKa = 4.19VNEE138 pKa = 4.47EE139 pKa = 4.01DD140 pKa = 4.53SSITFTIEE148 pKa = 5.8DD149 pKa = 3.7DD150 pKa = 4.51VKK152 pKa = 9.47WHH154 pKa = 7.13DD155 pKa = 3.9GEE157 pKa = 4.63EE158 pKa = 4.22VTAEE162 pKa = 3.79DD163 pKa = 2.96WAYY166 pKa = 9.07SYY168 pKa = 10.94EE169 pKa = 4.56VIMHH173 pKa = 7.09PDD175 pKa = 3.17YY176 pKa = 11.6DD177 pKa = 4.06GVRR180 pKa = 11.84GSSAGFAYY188 pKa = 10.74LEE190 pKa = 4.47GAQEE194 pKa = 4.17YY195 pKa = 10.0KK196 pKa = 10.74AGEE199 pKa = 4.16ADD201 pKa = 3.94SISGIEE207 pKa = 3.97IEE209 pKa = 4.38GEE211 pKa = 3.77KK212 pKa = 10.23TITFNYY218 pKa = 9.54TEE220 pKa = 4.64LTPSLLTGGVWGYY233 pKa = 10.41PMAKK237 pKa = 9.57HH238 pKa = 5.58VFEE241 pKa = 6.08DD242 pKa = 3.92IPVEE246 pKa = 4.24DD247 pKa = 4.18MASSPEE253 pKa = 3.95VRR255 pKa = 11.84EE256 pKa = 3.99EE257 pKa = 4.33PIGFGPYY264 pKa = 9.31KK265 pKa = 10.41VEE267 pKa = 3.79NVVPGEE273 pKa = 4.07SVTYY277 pKa = 10.73SKK279 pKa = 11.3FEE281 pKa = 4.61DD282 pKa = 3.82YY283 pKa = 10.28WRR285 pKa = 11.84GEE287 pKa = 4.06PNLDD291 pKa = 3.25EE292 pKa = 4.37VTLKK296 pKa = 10.7VVSPSNVANALEE308 pKa = 4.4TGDD311 pKa = 3.83VDD313 pKa = 5.66LVDD316 pKa = 5.12SFPADD321 pKa = 3.16QVVDD325 pKa = 4.41NIDD328 pKa = 3.55MPNVQWLGMTDD339 pKa = 3.59LAYY342 pKa = 9.64TYY344 pKa = 10.46IGFKK348 pKa = 10.63LGDD351 pKa = 3.36WNADD355 pKa = 3.41EE356 pKa = 6.22GEE358 pKa = 4.27VNYY361 pKa = 10.89KK362 pKa = 9.83GDD364 pKa = 3.76EE365 pKa = 3.99MKK367 pKa = 10.54MGDD370 pKa = 3.45KK371 pKa = 10.46KK372 pKa = 10.76LRR374 pKa = 11.84KK375 pKa = 9.43AMWYY379 pKa = 10.12AVDD382 pKa = 3.76NEE384 pKa = 4.83TVGEE388 pKa = 4.03RR389 pKa = 11.84FYY391 pKa = 11.8NGLRR395 pKa = 11.84WNGTTLIPPSHH406 pKa = 6.93PAWHH410 pKa = 7.62DD411 pKa = 3.59DD412 pKa = 3.91SIEE415 pKa = 4.12TPTYY419 pKa = 11.03DD420 pKa = 3.32EE421 pKa = 5.35DD422 pKa = 4.35KK423 pKa = 11.33ANEE426 pKa = 4.09LLDD429 pKa = 4.01DD430 pKa = 5.02AGYY433 pKa = 11.18KK434 pKa = 8.79DD435 pKa = 4.26TNDD438 pKa = 3.23DD439 pKa = 4.23GYY441 pKa = 11.32RR442 pKa = 11.84EE443 pKa = 4.22TPDD446 pKa = 3.85GEE448 pKa = 3.97EE449 pKa = 3.83LVINFASMDD458 pKa = 3.74SSDD461 pKa = 4.53LSEE464 pKa = 4.54PLANYY469 pKa = 8.64YY470 pKa = 8.97IQAWDD475 pKa = 3.52KK476 pKa = 11.58VGLNVQKK483 pKa = 10.93LDD485 pKa = 3.75GRR487 pKa = 11.84LHH489 pKa = 6.38EE490 pKa = 5.13FNSFYY495 pKa = 11.28DD496 pKa = 3.53KK497 pKa = 11.09VGQTGEE503 pKa = 4.45DD504 pKa = 3.8DD505 pKa = 4.21PNVDD509 pKa = 3.14VFMGAWSVGYY519 pKa = 10.32DD520 pKa = 3.43VDD522 pKa = 4.11PSGLYY527 pKa = 10.68GRR529 pKa = 11.84DD530 pKa = 2.84ALFNFTRR537 pKa = 11.84WANEE541 pKa = 3.78EE542 pKa = 3.85NDD544 pKa = 3.51EE545 pKa = 4.4LLAEE549 pKa = 4.61GLSEE553 pKa = 3.92EE554 pKa = 5.02AFDD557 pKa = 3.5TEE559 pKa = 4.21YY560 pKa = 11.1RR561 pKa = 11.84QDD563 pKa = 5.0VYY565 pKa = 11.22SQWQEE570 pKa = 3.7LMVEE574 pKa = 4.39EE575 pKa = 4.74VPVFPTLYY583 pKa = 9.46RR584 pKa = 11.84AMMVPANEE592 pKa = 3.76RR593 pKa = 11.84VVNYY597 pKa = 9.63DD598 pKa = 2.76ISVGSGVYY606 pKa = 10.09LYY608 pKa = 10.62EE609 pKa = 5.06LGVTQEE615 pKa = 4.21EE616 pKa = 4.86PVVEE620 pKa = 4.24
Molecular weight: 69.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N5BZQ6|A0A3N5BZQ6_9BACI CubicO group peptidase (Beta-lactamase class C family) OS=Aquisalibacillus elongatus OX=485577 GN=EDC24_0179 PE=4 SV=1
MM1 pKa = 7.52MKK3 pKa = 9.41RR4 pKa = 11.84TYY6 pKa = 10.32QPNKK10 pKa = 8.19RR11 pKa = 11.84KK12 pKa = 9.74RR13 pKa = 11.84NKK15 pKa = 8.71VHH17 pKa = 6.87GFRR20 pKa = 11.84TRR22 pKa = 11.84MKK24 pKa = 8.87TKK26 pKa = 10.27NGRR29 pKa = 11.84KK30 pKa = 8.66VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.2KK38 pKa = 10.05GRR40 pKa = 11.84KK41 pKa = 8.66VLSAA45 pKa = 4.05
MM1 pKa = 7.52MKK3 pKa = 9.41RR4 pKa = 11.84TYY6 pKa = 10.32QPNKK10 pKa = 8.19RR11 pKa = 11.84KK12 pKa = 9.74RR13 pKa = 11.84NKK15 pKa = 8.71VHH17 pKa = 6.87GFRR20 pKa = 11.84TRR22 pKa = 11.84MKK24 pKa = 8.87TKK26 pKa = 10.27NGRR29 pKa = 11.84KK30 pKa = 8.66VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.2KK38 pKa = 10.05GRR40 pKa = 11.84KK41 pKa = 8.66VLSAA45 pKa = 4.05
Molecular weight: 5.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
843163 |
29 |
2845 |
285.0 |
32.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.098 ± 0.049 | 0.58 ± 0.012 |
5.982 ± 0.042 | 8.122 ± 0.055 |
4.529 ± 0.041 | 6.599 ± 0.047 |
2.247 ± 0.021 | 7.762 ± 0.052 |
6.264 ± 0.041 | 9.585 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.768 ± 0.02 | 4.685 ± 0.036 |
3.481 ± 0.025 | 4.316 ± 0.039 |
3.903 ± 0.031 | 5.94 ± 0.039 |
5.243 ± 0.032 | 7.163 ± 0.04 |
0.997 ± 0.017 | 3.737 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
