
Ruminococcaceae bacterium TF06-43
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; unclassified Oscillospiraceae
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
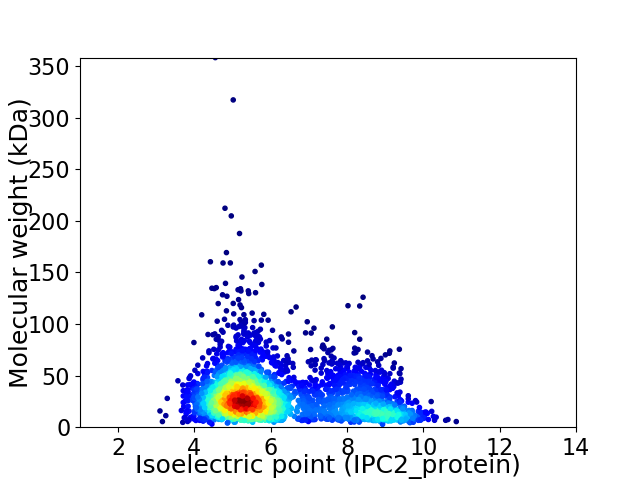
Virtual 2D-PAGE plot for 3276 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A417ND16|A0A417ND16_9FIRM Phosphatidic acid phosphatase OS=Ruminococcaceae bacterium TF06-43 OX=2292270 GN=DXC62_07160 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 10.41KK3 pKa = 10.59KK4 pKa = 10.24FMCMLMAAAMTLSMAACGSKK24 pKa = 10.42SDD26 pKa = 5.26DD27 pKa = 3.78TNTDD31 pKa = 3.46DD32 pKa = 4.49NAGGASDD39 pKa = 4.58GVQTFTVGTSGPLTGDD55 pKa = 3.18NAIYY59 pKa = 10.61GMAVKK64 pKa = 10.28QGVEE68 pKa = 3.78LAVNEE73 pKa = 4.34INASDD78 pKa = 3.67STIKK82 pKa = 10.81FEE84 pKa = 5.85FLSQDD89 pKa = 3.66DD90 pKa = 4.0EE91 pKa = 6.31ADD93 pKa = 3.5GEE95 pKa = 4.57KK96 pKa = 10.58AVNAYY101 pKa = 10.75NNMMDD106 pKa = 3.72NGMQVLVGPTTTGASIAVADD126 pKa = 3.61ACYY129 pKa = 10.06TDD131 pKa = 3.8RR132 pKa = 11.84TFMLTPSASSTDD144 pKa = 3.36VTAGKK149 pKa = 10.63DD150 pKa = 3.28NVFQVCFTDD159 pKa = 3.85PNQGVGAADD168 pKa = 3.3YY169 pKa = 9.06MAEE172 pKa = 4.02NFADD176 pKa = 3.53AKK178 pKa = 10.42IAVIYY183 pKa = 10.66RR184 pKa = 11.84NDD186 pKa = 3.46DD187 pKa = 3.57AYY189 pKa = 11.61SQGIRR194 pKa = 11.84DD195 pKa = 4.12AFVKK199 pKa = 10.43EE200 pKa = 4.33AGDD203 pKa = 3.52KK204 pKa = 10.78GLSIVYY210 pKa = 10.02QGTFTLDD217 pKa = 2.97TSSNFSVQLAAAQTAGADD235 pKa = 3.83LVFLPIYY242 pKa = 7.46YY243 pKa = 10.04QPASVIFAQAKK254 pKa = 9.03AMGYY258 pKa = 10.1APTFFGVDD266 pKa = 3.36GMDD269 pKa = 5.55GILDD273 pKa = 3.61MPGFDD278 pKa = 3.18TTLAEE283 pKa = 4.54GLVLMTPFCATVEE296 pKa = 4.26SSKK299 pKa = 11.29SFVDD303 pKa = 4.48AYY305 pKa = 8.85TAAYY309 pKa = 7.19GTTPNQFAADD319 pKa = 4.11AYY321 pKa = 10.39DD322 pKa = 3.07GVYY325 pKa = 9.66IVKK328 pKa = 10.28AALEE332 pKa = 4.18QAGCTADD339 pKa = 3.37MSNEE343 pKa = 4.19EE344 pKa = 3.87ICDD347 pKa = 3.81ALVSAITSLKK357 pKa = 9.31FTGLTGTDD365 pKa = 3.45MSWNAEE371 pKa = 4.09GQVSKK376 pKa = 11.26APTAYY381 pKa = 9.84VVKK384 pKa = 10.38DD385 pKa = 3.48GQYY388 pKa = 10.46VEE390 pKa = 4.63GG391 pKa = 4.4
MM1 pKa = 7.45KK2 pKa = 10.41KK3 pKa = 10.59KK4 pKa = 10.24FMCMLMAAAMTLSMAACGSKK24 pKa = 10.42SDD26 pKa = 5.26DD27 pKa = 3.78TNTDD31 pKa = 3.46DD32 pKa = 4.49NAGGASDD39 pKa = 4.58GVQTFTVGTSGPLTGDD55 pKa = 3.18NAIYY59 pKa = 10.61GMAVKK64 pKa = 10.28QGVEE68 pKa = 3.78LAVNEE73 pKa = 4.34INASDD78 pKa = 3.67STIKK82 pKa = 10.81FEE84 pKa = 5.85FLSQDD89 pKa = 3.66DD90 pKa = 4.0EE91 pKa = 6.31ADD93 pKa = 3.5GEE95 pKa = 4.57KK96 pKa = 10.58AVNAYY101 pKa = 10.75NNMMDD106 pKa = 3.72NGMQVLVGPTTTGASIAVADD126 pKa = 3.61ACYY129 pKa = 10.06TDD131 pKa = 3.8RR132 pKa = 11.84TFMLTPSASSTDD144 pKa = 3.36VTAGKK149 pKa = 10.63DD150 pKa = 3.28NVFQVCFTDD159 pKa = 3.85PNQGVGAADD168 pKa = 3.3YY169 pKa = 9.06MAEE172 pKa = 4.02NFADD176 pKa = 3.53AKK178 pKa = 10.42IAVIYY183 pKa = 10.66RR184 pKa = 11.84NDD186 pKa = 3.46DD187 pKa = 3.57AYY189 pKa = 11.61SQGIRR194 pKa = 11.84DD195 pKa = 4.12AFVKK199 pKa = 10.43EE200 pKa = 4.33AGDD203 pKa = 3.52KK204 pKa = 10.78GLSIVYY210 pKa = 10.02QGTFTLDD217 pKa = 2.97TSSNFSVQLAAAQTAGADD235 pKa = 3.83LVFLPIYY242 pKa = 7.46YY243 pKa = 10.04QPASVIFAQAKK254 pKa = 9.03AMGYY258 pKa = 10.1APTFFGVDD266 pKa = 3.36GMDD269 pKa = 5.55GILDD273 pKa = 3.61MPGFDD278 pKa = 3.18TTLAEE283 pKa = 4.54GLVLMTPFCATVEE296 pKa = 4.26SSKK299 pKa = 11.29SFVDD303 pKa = 4.48AYY305 pKa = 8.85TAAYY309 pKa = 7.19GTTPNQFAADD319 pKa = 4.11AYY321 pKa = 10.39DD322 pKa = 3.07GVYY325 pKa = 9.66IVKK328 pKa = 10.28AALEE332 pKa = 4.18QAGCTADD339 pKa = 3.37MSNEE343 pKa = 4.19EE344 pKa = 3.87ICDD347 pKa = 3.81ALVSAITSLKK357 pKa = 9.31FTGLTGTDD365 pKa = 3.45MSWNAEE371 pKa = 4.09GQVSKK376 pKa = 11.26APTAYY381 pKa = 9.84VVKK384 pKa = 10.38DD385 pKa = 3.48GQYY388 pKa = 10.46VEE390 pKa = 4.63GG391 pKa = 4.4
Molecular weight: 40.98 kDa
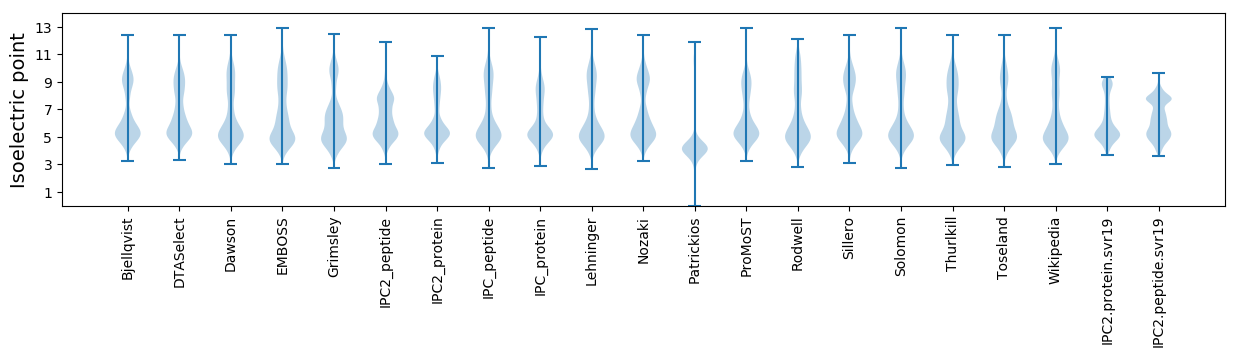
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A417MZ85|A0A417MZ85_9FIRM ATPase (Fragment) OS=Ruminococcaceae bacterium TF06-43 OX=2292270 GN=DXC62_16635 PE=4 SV=1
MM1 pKa = 7.53PRR3 pKa = 11.84MSKK6 pKa = 9.9KK7 pKa = 10.0RR8 pKa = 11.84RR9 pKa = 11.84LEE11 pKa = 3.44WSFFLRR17 pKa = 11.84QVKK20 pKa = 9.89VGNTTCDD27 pKa = 3.22RR28 pKa = 11.84ITYY31 pKa = 10.44NDD33 pKa = 3.57LCRR36 pKa = 11.84GCTHH40 pKa = 6.4SCKK43 pKa = 10.03QSFRR47 pKa = 11.84AVIILCPRR55 pKa = 11.84YY56 pKa = 8.96YY57 pKa = 10.38SKK59 pKa = 10.69RR60 pKa = 11.84RR61 pKa = 11.84KK62 pKa = 10.02KK63 pKa = 10.07EE64 pKa = 3.49DD65 pKa = 3.47RR66 pKa = 11.84DD67 pKa = 3.51NGRR70 pKa = 3.64
MM1 pKa = 7.53PRR3 pKa = 11.84MSKK6 pKa = 9.9KK7 pKa = 10.0RR8 pKa = 11.84RR9 pKa = 11.84LEE11 pKa = 3.44WSFFLRR17 pKa = 11.84QVKK20 pKa = 9.89VGNTTCDD27 pKa = 3.22RR28 pKa = 11.84ITYY31 pKa = 10.44NDD33 pKa = 3.57LCRR36 pKa = 11.84GCTHH40 pKa = 6.4SCKK43 pKa = 10.03QSFRR47 pKa = 11.84AVIILCPRR55 pKa = 11.84YY56 pKa = 8.96YY57 pKa = 10.38SKK59 pKa = 10.69RR60 pKa = 11.84RR61 pKa = 11.84KK62 pKa = 10.02KK63 pKa = 10.07EE64 pKa = 3.49DD65 pKa = 3.47RR66 pKa = 11.84DD67 pKa = 3.51NGRR70 pKa = 3.64
Molecular weight: 8.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
943759 |
23 |
3350 |
288.1 |
32.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.283 ± 0.048 | 1.722 ± 0.019 |
5.743 ± 0.036 | 7.037 ± 0.046 |
3.874 ± 0.029 | 7.201 ± 0.04 |
1.936 ± 0.023 | 5.914 ± 0.043 |
5.553 ± 0.042 | 9.659 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.813 ± 0.018 | 3.589 ± 0.027 |
3.749 ± 0.022 | 3.323 ± 0.024 |
5.544 ± 0.047 | 5.875 ± 0.033 |
5.633 ± 0.051 | 6.818 ± 0.033 |
0.993 ± 0.015 | 3.741 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
