
Pediococcus claussenii (strain ATCC BAA-344 / DSM 14800 / JCM 18046 / KCTC 3811 / P06)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Pediococcus; Pediococcus claussenii
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
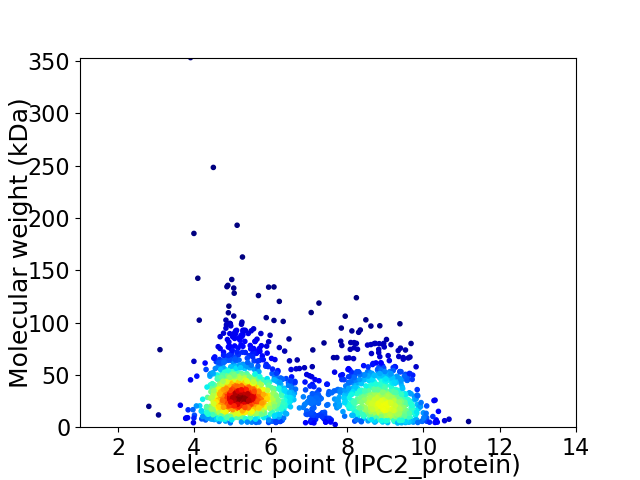
Virtual 2D-PAGE plot for 1877 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8PCH7|G8PCH7_PEDCP Putative membrane protein insertion efficiency factor OS=Pediococcus claussenii (strain ATCC BAA-344 / DSM 14800 / JCM 18046 / KCTC 3811 / P06) OX=701521 GN=PECL_670 PE=3 SV=1
MM1 pKa = 7.42SFWRR5 pKa = 11.84KK6 pKa = 7.2NHH8 pKa = 6.45KK9 pKa = 8.84STNLNTKK16 pKa = 7.89NHH18 pKa = 5.34YY19 pKa = 10.85KK20 pKa = 9.61MYY22 pKa = 10.47KK23 pKa = 10.0AGKK26 pKa = 7.78QWLFAGITTIAGLGGQSIAGAKK48 pKa = 9.79ADD50 pKa = 4.08SLSATSTATKK60 pKa = 10.18DD61 pKa = 3.6VQDD64 pKa = 3.8QDD66 pKa = 3.8VLATGDD72 pKa = 3.67SAIIPAEE79 pKa = 3.99SSVASSSVATASATSATLSSASSAVSSSNSSNTSSASASSADD121 pKa = 3.68TSSSSSSVASGSNTTSSSVSSTSSSSATSSANASSNNSNSSSATSSTVNSSSNSAATSSTAEE183 pKa = 3.87SSSPVSSSSSVASITSNALSSATSGTQSLVSSGVTNTTSSSATTSSTATSSSAVASLSMSNSMSLSEE250 pKa = 4.14SQSLISLASLNIGSSVAASSAASSAKK276 pKa = 9.35KK277 pKa = 10.09VAMKK281 pKa = 10.74AMVMAATVAPTVAVNVGSLSYY302 pKa = 11.86NMGTQIAYY310 pKa = 9.56YY311 pKa = 10.64ALRR314 pKa = 11.84SWKK317 pKa = 10.37DD318 pKa = 2.94SGLNIVSLTPSWLLANFTAISALLTYY344 pKa = 9.26YY345 pKa = 10.01QANGTVLRR353 pKa = 11.84ADD355 pKa = 3.46GTMDD359 pKa = 4.0PSLIAALSGIYY370 pKa = 9.58AAPSWSGQTGIKK382 pKa = 10.38VSGSTAASSGYY393 pKa = 6.49TQNGSGWNNTDD404 pKa = 3.13YY405 pKa = 11.62TNDD408 pKa = 3.25YY409 pKa = 10.34YY410 pKa = 11.59LIGTVKK416 pKa = 10.64NPFSATDD423 pKa = 3.62YY424 pKa = 9.59YY425 pKa = 11.65AGIASILNVYY435 pKa = 8.78AQEE438 pKa = 3.93VTNYY442 pKa = 9.25FDD444 pKa = 3.63QVATKK449 pKa = 10.6VQDD452 pKa = 3.35TSGGVGNSLSDD463 pKa = 3.35QLASYY468 pKa = 7.49FTYY471 pKa = 10.78NPNIAQNQSGALGLLANVTALVSPLLSAQGSNDD504 pKa = 3.31SLTGQIPGIFGFHH517 pKa = 5.56VTTSDD522 pKa = 3.52PLIISDD528 pKa = 4.3LTQNASNAYY537 pKa = 8.15NTGSLQSLDD546 pKa = 3.64NLISVVMSSLQPAIEE561 pKa = 4.02KK562 pKa = 10.55QLFNDD567 pKa = 3.28IRR569 pKa = 11.84TLGINQSGSLSIPSDD584 pKa = 3.92LYY586 pKa = 9.14TAVYY590 pKa = 9.44TLTPGLAQVISYY602 pKa = 10.01FNFSSLLNLPLLNQIYY618 pKa = 10.22LSVVNGIRR626 pKa = 11.84TAIVNDD632 pKa = 4.58AIVGVNAAFKK642 pKa = 10.86NILDD646 pKa = 4.52PKK648 pKa = 10.57DD649 pKa = 4.28NPDD652 pKa = 3.58PDD654 pKa = 3.86YY655 pKa = 11.21STVAGYY661 pKa = 10.78NRR663 pKa = 11.84TSASGAANTVQTTQNTTLLAQSVGYY688 pKa = 9.95SWAQQVVPTILRR700 pKa = 11.84AASADD705 pKa = 3.67ALNKK709 pKa = 8.25ATPQNLNEE717 pKa = 4.41ILNTLLNAGVITSAQVTQLTVAGTQNTSYY746 pKa = 10.07TNYY749 pKa = 10.06VADD752 pKa = 3.58PTGAQNLITSLYY764 pKa = 7.23TAEE767 pKa = 4.1YY768 pKa = 10.35NAVQQAVTDD777 pKa = 4.26FNNGTPKK784 pKa = 10.48GANPTWTDD792 pKa = 3.15PSTVQTDD799 pKa = 3.75GTNSGIEE806 pKa = 3.97TLTDD810 pKa = 3.13YY811 pKa = 11.81GNVYY815 pKa = 10.64DD816 pKa = 4.14YY817 pKa = 11.15LVKK820 pKa = 10.39VRR822 pKa = 11.84HH823 pKa = 6.33DD824 pKa = 3.72AMVDD828 pKa = 3.14ADD830 pKa = 4.11AKK832 pKa = 10.64SHH834 pKa = 5.49SEE836 pKa = 3.78KK837 pKa = 10.48DD838 pKa = 3.48GKK840 pKa = 10.39VAALTSVSGSTNANFNTTAGLTNYY864 pKa = 7.09VTNAVSASGILSSTSDD880 pKa = 3.01ISALAAISQTDD891 pKa = 3.66YY892 pKa = 10.47VSEE895 pKa = 4.37YY896 pKa = 10.1NAEE899 pKa = 3.97AALTNTAFAQGAAAAQVQIRR919 pKa = 11.84LHH921 pKa = 6.3PNGHH925 pKa = 6.54LASPASVDD933 pKa = 3.38YY934 pKa = 10.94NGYY937 pKa = 10.02SGSSDD942 pKa = 2.71KK943 pKa = 10.44TAQYY947 pKa = 11.62AVTTDD952 pKa = 3.42TLQTGQAFTDD962 pKa = 5.04GYY964 pKa = 9.82LASVDD969 pKa = 3.86TVTEE973 pKa = 4.03IAAPSAPTSSWTTAQLTAFNTAIKK997 pKa = 10.49NAFPNATQQSNGNWVIGTDD1016 pKa = 3.92YY1017 pKa = 10.77VAPGATNVLYY1027 pKa = 8.0TAPTTVTGFGSTNQTLTTAGAAGNTSDD1054 pKa = 3.44TVTFTFTAGVNLISNYY1070 pKa = 10.43GIDD1073 pKa = 4.06GASSTGLGQDD1083 pKa = 3.23TSFTPTGTALTIPGLLTGLNGEE1105 pKa = 4.17LQTWHH1110 pKa = 6.57NASKK1114 pKa = 10.69ILFDD1118 pKa = 4.42FNNTTFSTTSNGVTGSSLSIYY1139 pKa = 10.27LLAYY1143 pKa = 8.94YY1144 pKa = 10.02AQLGYY1149 pKa = 9.33ATATDD1154 pKa = 4.1LQTAISNEE1162 pKa = 3.95FASGSDD1168 pKa = 3.2KK1169 pKa = 11.17LALQVLALIQGGLATTSAASSGTVTMNYY1197 pKa = 10.2AFKK1200 pKa = 9.54NTNVNSTVATVGSIVDD1216 pKa = 3.33QRR1218 pKa = 11.84GNKK1221 pKa = 8.45LQPDD1225 pKa = 4.07TTNSLVVGNNLVFQTPTIPTGRR1247 pKa = 11.84TINFDD1252 pKa = 3.04NSYY1255 pKa = 10.85VQMSLDD1261 pKa = 2.97GSEE1264 pKa = 4.05PVKK1267 pKa = 10.41YY1268 pKa = 8.89TLTQLTQILGITLTGNSQADD1288 pKa = 3.66LTSIINAFNEE1298 pKa = 3.92ANGNVTITTGNLLDD1312 pKa = 4.08NLQALTYY1319 pKa = 10.14IQLNPQASTASVSGSQSVTYY1339 pKa = 9.36TGSVPALTTGDD1350 pKa = 3.89YY1351 pKa = 10.35TVTLADD1357 pKa = 3.6GSTYY1361 pKa = 10.16TLQAGDD1367 pKa = 3.44IDD1369 pKa = 4.95IINSSKK1375 pKa = 10.86NAGTYY1380 pKa = 7.73QVEE1383 pKa = 4.36LTAQGLANINAASADD1398 pKa = 3.92HH1399 pKa = 7.3IYY1401 pKa = 10.61TIDD1404 pKa = 3.56NSTLGTLVINKK1415 pKa = 9.51ANATINVDD1423 pKa = 3.66GTQHH1427 pKa = 6.48VYY1429 pKa = 10.01TGDD1432 pKa = 3.67TYY1434 pKa = 11.7NATATATGAVNGEE1447 pKa = 4.18TLNYY1451 pKa = 8.21TVSGSQTEE1459 pKa = 4.37IGSSPVAVNFDD1470 pKa = 3.62PADD1473 pKa = 4.72PINANYY1479 pKa = 10.39NITVTGAAVITVVRR1493 pKa = 11.84NATSDD1498 pKa = 3.81SNSTSASEE1506 pKa = 4.22SMSDD1510 pKa = 3.55LGSVSASDD1518 pKa = 3.37ITSVSQSNSIAGSISISNSNNFSASNSIVEE1548 pKa = 4.45SASTSASKK1556 pKa = 10.58QSSIEE1561 pKa = 4.16TNDD1564 pKa = 3.34KK1565 pKa = 10.75VSEE1568 pKa = 4.08LMSDD1572 pKa = 3.48SASLSEE1578 pKa = 4.06EE1579 pKa = 3.68ASAGGSQSLQNSQSLSNSMSTNGDD1603 pKa = 3.1NSISNVNSLSEE1614 pKa = 4.44SNSLSAQNSLSIAQGNSLNNEE1635 pKa = 3.85LSDD1638 pKa = 4.07AEE1640 pKa = 4.39SGVASGDD1647 pKa = 3.54TSQSNSVNDD1656 pKa = 4.16AGSQSLVNSVSEE1668 pKa = 4.41SNSIIDD1674 pKa = 3.78RR1675 pKa = 11.84GSDD1678 pKa = 3.28GSNDD1682 pKa = 3.91SISNSNSYY1690 pKa = 10.86SVANSNSVQNSISASNSLSLDD1711 pKa = 3.16DD1712 pKa = 4.65SNNVNSDD1719 pKa = 2.94TSEE1722 pKa = 4.19SVSLSISNSQNIVNSVSTSTSGSTSFSMSGNEE1754 pKa = 4.11SVSGSTSISEE1764 pKa = 4.35SEE1766 pKa = 4.29SLSSVDD1772 pKa = 3.63SVSGSVSNSNNVSNTTSASLSLSNSNSEE1800 pKa = 4.19STVEE1804 pKa = 4.09SLSVSNSLSGFAKK1817 pKa = 10.55CC1818 pKa = 5.43
MM1 pKa = 7.42SFWRR5 pKa = 11.84KK6 pKa = 7.2NHH8 pKa = 6.45KK9 pKa = 8.84STNLNTKK16 pKa = 7.89NHH18 pKa = 5.34YY19 pKa = 10.85KK20 pKa = 9.61MYY22 pKa = 10.47KK23 pKa = 10.0AGKK26 pKa = 7.78QWLFAGITTIAGLGGQSIAGAKK48 pKa = 9.79ADD50 pKa = 4.08SLSATSTATKK60 pKa = 10.18DD61 pKa = 3.6VQDD64 pKa = 3.8QDD66 pKa = 3.8VLATGDD72 pKa = 3.67SAIIPAEE79 pKa = 3.99SSVASSSVATASATSATLSSASSAVSSSNSSNTSSASASSADD121 pKa = 3.68TSSSSSSVASGSNTTSSSVSSTSSSSATSSANASSNNSNSSSATSSTVNSSSNSAATSSTAEE183 pKa = 3.87SSSPVSSSSSVASITSNALSSATSGTQSLVSSGVTNTTSSSATTSSTATSSSAVASLSMSNSMSLSEE250 pKa = 4.14SQSLISLASLNIGSSVAASSAASSAKK276 pKa = 9.35KK277 pKa = 10.09VAMKK281 pKa = 10.74AMVMAATVAPTVAVNVGSLSYY302 pKa = 11.86NMGTQIAYY310 pKa = 9.56YY311 pKa = 10.64ALRR314 pKa = 11.84SWKK317 pKa = 10.37DD318 pKa = 2.94SGLNIVSLTPSWLLANFTAISALLTYY344 pKa = 9.26YY345 pKa = 10.01QANGTVLRR353 pKa = 11.84ADD355 pKa = 3.46GTMDD359 pKa = 4.0PSLIAALSGIYY370 pKa = 9.58AAPSWSGQTGIKK382 pKa = 10.38VSGSTAASSGYY393 pKa = 6.49TQNGSGWNNTDD404 pKa = 3.13YY405 pKa = 11.62TNDD408 pKa = 3.25YY409 pKa = 10.34YY410 pKa = 11.59LIGTVKK416 pKa = 10.64NPFSATDD423 pKa = 3.62YY424 pKa = 9.59YY425 pKa = 11.65AGIASILNVYY435 pKa = 8.78AQEE438 pKa = 3.93VTNYY442 pKa = 9.25FDD444 pKa = 3.63QVATKK449 pKa = 10.6VQDD452 pKa = 3.35TSGGVGNSLSDD463 pKa = 3.35QLASYY468 pKa = 7.49FTYY471 pKa = 10.78NPNIAQNQSGALGLLANVTALVSPLLSAQGSNDD504 pKa = 3.31SLTGQIPGIFGFHH517 pKa = 5.56VTTSDD522 pKa = 3.52PLIISDD528 pKa = 4.3LTQNASNAYY537 pKa = 8.15NTGSLQSLDD546 pKa = 3.64NLISVVMSSLQPAIEE561 pKa = 4.02KK562 pKa = 10.55QLFNDD567 pKa = 3.28IRR569 pKa = 11.84TLGINQSGSLSIPSDD584 pKa = 3.92LYY586 pKa = 9.14TAVYY590 pKa = 9.44TLTPGLAQVISYY602 pKa = 10.01FNFSSLLNLPLLNQIYY618 pKa = 10.22LSVVNGIRR626 pKa = 11.84TAIVNDD632 pKa = 4.58AIVGVNAAFKK642 pKa = 10.86NILDD646 pKa = 4.52PKK648 pKa = 10.57DD649 pKa = 4.28NPDD652 pKa = 3.58PDD654 pKa = 3.86YY655 pKa = 11.21STVAGYY661 pKa = 10.78NRR663 pKa = 11.84TSASGAANTVQTTQNTTLLAQSVGYY688 pKa = 9.95SWAQQVVPTILRR700 pKa = 11.84AASADD705 pKa = 3.67ALNKK709 pKa = 8.25ATPQNLNEE717 pKa = 4.41ILNTLLNAGVITSAQVTQLTVAGTQNTSYY746 pKa = 10.07TNYY749 pKa = 10.06VADD752 pKa = 3.58PTGAQNLITSLYY764 pKa = 7.23TAEE767 pKa = 4.1YY768 pKa = 10.35NAVQQAVTDD777 pKa = 4.26FNNGTPKK784 pKa = 10.48GANPTWTDD792 pKa = 3.15PSTVQTDD799 pKa = 3.75GTNSGIEE806 pKa = 3.97TLTDD810 pKa = 3.13YY811 pKa = 11.81GNVYY815 pKa = 10.64DD816 pKa = 4.14YY817 pKa = 11.15LVKK820 pKa = 10.39VRR822 pKa = 11.84HH823 pKa = 6.33DD824 pKa = 3.72AMVDD828 pKa = 3.14ADD830 pKa = 4.11AKK832 pKa = 10.64SHH834 pKa = 5.49SEE836 pKa = 3.78KK837 pKa = 10.48DD838 pKa = 3.48GKK840 pKa = 10.39VAALTSVSGSTNANFNTTAGLTNYY864 pKa = 7.09VTNAVSASGILSSTSDD880 pKa = 3.01ISALAAISQTDD891 pKa = 3.66YY892 pKa = 10.47VSEE895 pKa = 4.37YY896 pKa = 10.1NAEE899 pKa = 3.97AALTNTAFAQGAAAAQVQIRR919 pKa = 11.84LHH921 pKa = 6.3PNGHH925 pKa = 6.54LASPASVDD933 pKa = 3.38YY934 pKa = 10.94NGYY937 pKa = 10.02SGSSDD942 pKa = 2.71KK943 pKa = 10.44TAQYY947 pKa = 11.62AVTTDD952 pKa = 3.42TLQTGQAFTDD962 pKa = 5.04GYY964 pKa = 9.82LASVDD969 pKa = 3.86TVTEE973 pKa = 4.03IAAPSAPTSSWTTAQLTAFNTAIKK997 pKa = 10.49NAFPNATQQSNGNWVIGTDD1016 pKa = 3.92YY1017 pKa = 10.77VAPGATNVLYY1027 pKa = 8.0TAPTTVTGFGSTNQTLTTAGAAGNTSDD1054 pKa = 3.44TVTFTFTAGVNLISNYY1070 pKa = 10.43GIDD1073 pKa = 4.06GASSTGLGQDD1083 pKa = 3.23TSFTPTGTALTIPGLLTGLNGEE1105 pKa = 4.17LQTWHH1110 pKa = 6.57NASKK1114 pKa = 10.69ILFDD1118 pKa = 4.42FNNTTFSTTSNGVTGSSLSIYY1139 pKa = 10.27LLAYY1143 pKa = 8.94YY1144 pKa = 10.02AQLGYY1149 pKa = 9.33ATATDD1154 pKa = 4.1LQTAISNEE1162 pKa = 3.95FASGSDD1168 pKa = 3.2KK1169 pKa = 11.17LALQVLALIQGGLATTSAASSGTVTMNYY1197 pKa = 10.2AFKK1200 pKa = 9.54NTNVNSTVATVGSIVDD1216 pKa = 3.33QRR1218 pKa = 11.84GNKK1221 pKa = 8.45LQPDD1225 pKa = 4.07TTNSLVVGNNLVFQTPTIPTGRR1247 pKa = 11.84TINFDD1252 pKa = 3.04NSYY1255 pKa = 10.85VQMSLDD1261 pKa = 2.97GSEE1264 pKa = 4.05PVKK1267 pKa = 10.41YY1268 pKa = 8.89TLTQLTQILGITLTGNSQADD1288 pKa = 3.66LTSIINAFNEE1298 pKa = 3.92ANGNVTITTGNLLDD1312 pKa = 4.08NLQALTYY1319 pKa = 10.14IQLNPQASTASVSGSQSVTYY1339 pKa = 9.36TGSVPALTTGDD1350 pKa = 3.89YY1351 pKa = 10.35TVTLADD1357 pKa = 3.6GSTYY1361 pKa = 10.16TLQAGDD1367 pKa = 3.44IDD1369 pKa = 4.95IINSSKK1375 pKa = 10.86NAGTYY1380 pKa = 7.73QVEE1383 pKa = 4.36LTAQGLANINAASADD1398 pKa = 3.92HH1399 pKa = 7.3IYY1401 pKa = 10.61TIDD1404 pKa = 3.56NSTLGTLVINKK1415 pKa = 9.51ANATINVDD1423 pKa = 3.66GTQHH1427 pKa = 6.48VYY1429 pKa = 10.01TGDD1432 pKa = 3.67TYY1434 pKa = 11.7NATATATGAVNGEE1447 pKa = 4.18TLNYY1451 pKa = 8.21TVSGSQTEE1459 pKa = 4.37IGSSPVAVNFDD1470 pKa = 3.62PADD1473 pKa = 4.72PINANYY1479 pKa = 10.39NITVTGAAVITVVRR1493 pKa = 11.84NATSDD1498 pKa = 3.81SNSTSASEE1506 pKa = 4.22SMSDD1510 pKa = 3.55LGSVSASDD1518 pKa = 3.37ITSVSQSNSIAGSISISNSNNFSASNSIVEE1548 pKa = 4.45SASTSASKK1556 pKa = 10.58QSSIEE1561 pKa = 4.16TNDD1564 pKa = 3.34KK1565 pKa = 10.75VSEE1568 pKa = 4.08LMSDD1572 pKa = 3.48SASLSEE1578 pKa = 4.06EE1579 pKa = 3.68ASAGGSQSLQNSQSLSNSMSTNGDD1603 pKa = 3.1NSISNVNSLSEE1614 pKa = 4.44SNSLSAQNSLSIAQGNSLNNEE1635 pKa = 3.85LSDD1638 pKa = 4.07AEE1640 pKa = 4.39SGVASGDD1647 pKa = 3.54TSQSNSVNDD1656 pKa = 4.16AGSQSLVNSVSEE1668 pKa = 4.41SNSIIDD1674 pKa = 3.78RR1675 pKa = 11.84GSDD1678 pKa = 3.28GSNDD1682 pKa = 3.91SISNSNSYY1690 pKa = 10.86SVANSNSVQNSISASNSLSLDD1711 pKa = 3.16DD1712 pKa = 4.65SNNVNSDD1719 pKa = 2.94TSEE1722 pKa = 4.19SVSLSISNSQNIVNSVSTSTSGSTSFSMSGNEE1754 pKa = 4.11SVSGSTSISEE1764 pKa = 4.35SEE1766 pKa = 4.29SLSSVDD1772 pKa = 3.63SVSGSVSNSNNVSNTTSASLSLSNSNSEE1800 pKa = 4.19STVEE1804 pKa = 4.09SLSVSNSLSGFAKK1817 pKa = 10.55CC1818 pKa = 5.43
Molecular weight: 185.2 kDa
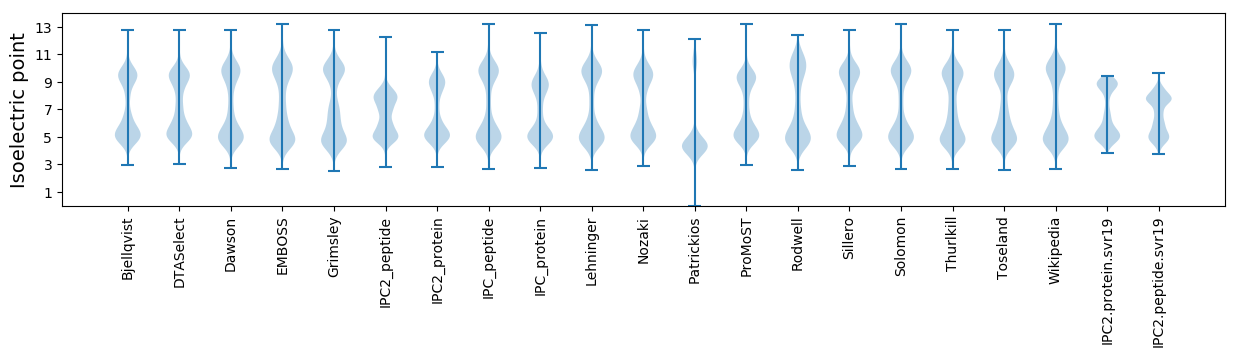
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8PE98|G8PE98_PEDCP NADH oxidase OS=Pediococcus claussenii (strain ATCC BAA-344 / DSM 14800 / JCM 18046 / KCTC 3811 / P06) OX=701521 GN=nox PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.69KK9 pKa = 7.77RR10 pKa = 11.84HH11 pKa = 4.99RR12 pKa = 11.84QRR14 pKa = 11.84VHH16 pKa = 6.07GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84RR29 pKa = 11.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.8VLSAA44 pKa = 4.11
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.69KK9 pKa = 7.77RR10 pKa = 11.84HH11 pKa = 4.99RR12 pKa = 11.84QRR14 pKa = 11.84VHH16 pKa = 6.07GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84RR29 pKa = 11.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84QRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.8VLSAA44 pKa = 4.11
Molecular weight: 5.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
566800 |
21 |
3609 |
302.0 |
33.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.024 ± 0.128 | 0.379 ± 0.011 |
5.874 ± 0.069 | 5.944 ± 0.078 |
4.496 ± 0.047 | 6.606 ± 0.055 |
1.878 ± 0.025 | 7.832 ± 0.073 |
6.809 ± 0.07 | 9.401 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.708 ± 0.032 | 5.541 ± 0.052 |
3.314 ± 0.031 | 4.232 ± 0.046 |
4.052 ± 0.056 | 6.698 ± 0.134 |
5.745 ± 0.063 | 7.146 ± 0.049 |
0.978 ± 0.021 | 3.344 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
