
Diaporthe helianthi
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Sordariomycetidae; Diaporthales; Diaporthaceae; Diaporthe
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
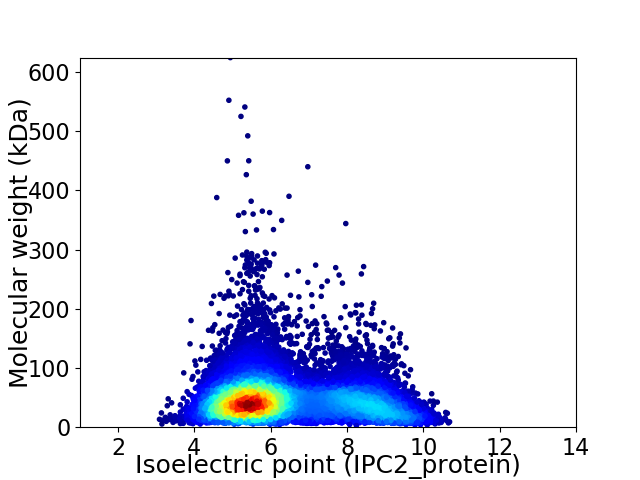
Virtual 2D-PAGE plot for 13137 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P5ICH9|A0A2P5ICH9_9PEZI TLP1_add_C domain-containing protein OS=Diaporthe helianthi OX=158607 GN=DHEL01_v201359 PE=4 SV=1
MM1 pKa = 7.38KK2 pKa = 10.34ACAEE6 pKa = 4.08PALLALLLAVTASAKK21 pKa = 9.22VVRR24 pKa = 11.84IPVARR29 pKa = 11.84NSPGATVSRR38 pKa = 11.84RR39 pKa = 11.84HH40 pKa = 5.97HH41 pKa = 6.43PRR43 pKa = 11.84LTPRR47 pKa = 11.84ASFTEE52 pKa = 4.07SLVNNITGGGYY63 pKa = 8.88YY64 pKa = 10.59ASVSVGTPGQDD75 pKa = 2.42IDD77 pKa = 4.59MILDD81 pKa = 3.81TGSSDD86 pKa = 3.54AFIVAAGADD95 pKa = 3.92LCQSPRR101 pKa = 11.84LQLIYY106 pKa = 9.38QTQCGQTFDD115 pKa = 4.26PSRR118 pKa = 11.84SSTFDD123 pKa = 3.07VVVPDD128 pKa = 4.37GFQIQYY134 pKa = 10.66LDD136 pKa = 3.53GSTASGDD143 pKa = 3.87YY144 pKa = 9.63ITDD147 pKa = 3.28HH148 pKa = 6.83FEE150 pKa = 4.58IGDD153 pKa = 3.92TVIEE157 pKa = 4.14SLQMGLANQTASGTGVLGIGYY178 pKa = 6.69TANEE182 pKa = 4.23AAEE185 pKa = 4.17TLYY188 pKa = 10.66PNLVDD193 pKa = 4.89QMVAQDD199 pKa = 4.83FIPTKK204 pKa = 10.5AYY206 pKa = 10.58SLYY209 pKa = 10.95LNDD212 pKa = 4.1YY213 pKa = 9.53QSSTGSILFGGVDD226 pKa = 3.45TEE228 pKa = 4.26KK229 pKa = 10.88FIGGLTVIPVLRR241 pKa = 11.84DD242 pKa = 3.15AQTQDD247 pKa = 3.19FTSFTVGMTAISYY260 pKa = 8.17AFPNGTTGDD269 pKa = 3.53ASLQGGSLDD278 pKa = 4.87AILDD282 pKa = 3.76SGTTLSYY289 pKa = 10.87FPDD292 pKa = 3.83SVATPLFEE300 pKa = 5.31ALGAFTYY307 pKa = 10.72SSLGSVGLTMVEE319 pKa = 4.27CSLATSGLEE328 pKa = 3.56LTFRR332 pKa = 11.84FNDD335 pKa = 3.47SVSIVVPADD344 pKa = 3.28EE345 pKa = 4.33VVIDD349 pKa = 3.92AFGPIEE355 pKa = 4.29GNIPPEE361 pKa = 4.39IPLEE365 pKa = 4.13QPCLFGIQNSGDD377 pKa = 3.8SLPSEE382 pKa = 4.75GARR385 pKa = 11.84QIDD388 pKa = 3.84FALLGDD394 pKa = 3.95TFLRR398 pKa = 11.84SAYY401 pKa = 9.94VVYY404 pKa = 10.81DD405 pKa = 3.9LDD407 pKa = 3.98NNEE410 pKa = 3.9IAMAQANLNSTDD422 pKa = 3.51TNVQEE427 pKa = 4.31LTEE430 pKa = 4.41SSDD433 pKa = 3.31LSSFEE438 pKa = 4.76GVEE441 pKa = 4.01QQQQSSGTSGGGSSGTDD458 pKa = 3.11SSSSAGSSGTLSQDD472 pKa = 3.39FGRR475 pKa = 11.84CEE477 pKa = 3.76FGLEE481 pKa = 3.87CGYY484 pKa = 9.99GQRR487 pKa = 11.84GRR489 pKa = 11.84GGDD492 pKa = 3.76CDD494 pKa = 5.19DD495 pKa = 6.16DD496 pKa = 3.66EE497 pKa = 4.78HH498 pKa = 8.29QRR500 pKa = 11.84DD501 pKa = 3.63LLFDD505 pKa = 4.54CGLGAVRR512 pKa = 11.84PPFTFGQCQQDD523 pKa = 3.41RR524 pKa = 11.84AQQQ527 pKa = 3.21
MM1 pKa = 7.38KK2 pKa = 10.34ACAEE6 pKa = 4.08PALLALLLAVTASAKK21 pKa = 9.22VVRR24 pKa = 11.84IPVARR29 pKa = 11.84NSPGATVSRR38 pKa = 11.84RR39 pKa = 11.84HH40 pKa = 5.97HH41 pKa = 6.43PRR43 pKa = 11.84LTPRR47 pKa = 11.84ASFTEE52 pKa = 4.07SLVNNITGGGYY63 pKa = 8.88YY64 pKa = 10.59ASVSVGTPGQDD75 pKa = 2.42IDD77 pKa = 4.59MILDD81 pKa = 3.81TGSSDD86 pKa = 3.54AFIVAAGADD95 pKa = 3.92LCQSPRR101 pKa = 11.84LQLIYY106 pKa = 9.38QTQCGQTFDD115 pKa = 4.26PSRR118 pKa = 11.84SSTFDD123 pKa = 3.07VVVPDD128 pKa = 4.37GFQIQYY134 pKa = 10.66LDD136 pKa = 3.53GSTASGDD143 pKa = 3.87YY144 pKa = 9.63ITDD147 pKa = 3.28HH148 pKa = 6.83FEE150 pKa = 4.58IGDD153 pKa = 3.92TVIEE157 pKa = 4.14SLQMGLANQTASGTGVLGIGYY178 pKa = 6.69TANEE182 pKa = 4.23AAEE185 pKa = 4.17TLYY188 pKa = 10.66PNLVDD193 pKa = 4.89QMVAQDD199 pKa = 4.83FIPTKK204 pKa = 10.5AYY206 pKa = 10.58SLYY209 pKa = 10.95LNDD212 pKa = 4.1YY213 pKa = 9.53QSSTGSILFGGVDD226 pKa = 3.45TEE228 pKa = 4.26KK229 pKa = 10.88FIGGLTVIPVLRR241 pKa = 11.84DD242 pKa = 3.15AQTQDD247 pKa = 3.19FTSFTVGMTAISYY260 pKa = 8.17AFPNGTTGDD269 pKa = 3.53ASLQGGSLDD278 pKa = 4.87AILDD282 pKa = 3.76SGTTLSYY289 pKa = 10.87FPDD292 pKa = 3.83SVATPLFEE300 pKa = 5.31ALGAFTYY307 pKa = 10.72SSLGSVGLTMVEE319 pKa = 4.27CSLATSGLEE328 pKa = 3.56LTFRR332 pKa = 11.84FNDD335 pKa = 3.47SVSIVVPADD344 pKa = 3.28EE345 pKa = 4.33VVIDD349 pKa = 3.92AFGPIEE355 pKa = 4.29GNIPPEE361 pKa = 4.39IPLEE365 pKa = 4.13QPCLFGIQNSGDD377 pKa = 3.8SLPSEE382 pKa = 4.75GARR385 pKa = 11.84QIDD388 pKa = 3.84FALLGDD394 pKa = 3.95TFLRR398 pKa = 11.84SAYY401 pKa = 9.94VVYY404 pKa = 10.81DD405 pKa = 3.9LDD407 pKa = 3.98NNEE410 pKa = 3.9IAMAQANLNSTDD422 pKa = 3.51TNVQEE427 pKa = 4.31LTEE430 pKa = 4.41SSDD433 pKa = 3.31LSSFEE438 pKa = 4.76GVEE441 pKa = 4.01QQQQSSGTSGGGSSGTDD458 pKa = 3.11SSSSAGSSGTLSQDD472 pKa = 3.39FGRR475 pKa = 11.84CEE477 pKa = 3.76FGLEE481 pKa = 3.87CGYY484 pKa = 9.99GQRR487 pKa = 11.84GRR489 pKa = 11.84GGDD492 pKa = 3.76CDD494 pKa = 5.19DD495 pKa = 6.16DD496 pKa = 3.66EE497 pKa = 4.78HH498 pKa = 8.29QRR500 pKa = 11.84DD501 pKa = 3.63LLFDD505 pKa = 4.54CGLGAVRR512 pKa = 11.84PPFTFGQCQQDD523 pKa = 3.41RR524 pKa = 11.84AQQQ527 pKa = 3.21
Molecular weight: 55.46 kDa
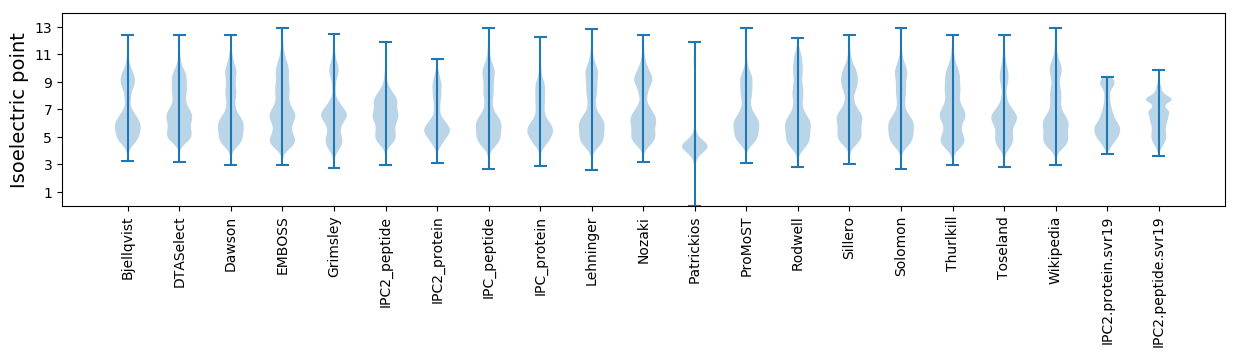
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P5HX61|A0A2P5HX61_9PEZI Uncharacterized protein OS=Diaporthe helianthi OX=158607 GN=DHEL01_v206758 PE=3 SV=1
MM1 pKa = 7.82DD2 pKa = 5.28SDD4 pKa = 4.27MTTMHH9 pKa = 6.94HH10 pKa = 6.22SLSGSSQCPATLASPQLLARR30 pKa = 11.84SVDD33 pKa = 3.76AILRR37 pKa = 11.84PDD39 pKa = 3.39HH40 pKa = 6.58VFSLSRR46 pKa = 11.84LVSISLPGIYY56 pKa = 9.21TRR58 pKa = 11.84PHH60 pKa = 5.27TKK62 pKa = 9.96GVNGVGLGPWYY73 pKa = 10.32RR74 pKa = 11.84AVLGLLPPNYY84 pKa = 9.87EE85 pKa = 3.95NLLCRR90 pKa = 11.84RR91 pKa = 11.84LGVQSIWASPIDD103 pKa = 4.27LLQPPAASVRR113 pKa = 11.84VCSKK117 pKa = 10.15ISTSGSFPAADD128 pKa = 3.9FRR130 pKa = 11.84IALIGRR136 pKa = 11.84MPQQLFVHH144 pKa = 6.67LSSAPRR150 pKa = 11.84CHH152 pKa = 6.92DD153 pKa = 3.23VALRR157 pKa = 11.84RR158 pKa = 11.84SPKK161 pKa = 10.04LIMRR165 pKa = 11.84STHH168 pKa = 6.28PPGSFCAPNLQARR181 pKa = 11.84SMWASWAAGHH191 pKa = 5.95QRR193 pKa = 11.84VNHH196 pKa = 6.1GLPLATWGLGQRR208 pKa = 11.84LKK210 pKa = 10.01TLPGDD215 pKa = 3.11SRR217 pKa = 11.84YY218 pKa = 9.54RR219 pKa = 11.84VSRR222 pKa = 11.84AGFTGPHH229 pKa = 5.98KK230 pKa = 10.72CWLTGMAKK238 pKa = 10.12KK239 pKa = 9.91RR240 pKa = 11.84DD241 pKa = 3.88KK242 pKa = 9.93PTEE245 pKa = 3.68LAEE248 pKa = 4.05QGRR251 pKa = 11.84LQIPRR256 pKa = 11.84TLSDD260 pKa = 3.54LGGSSSQNPALIGRR274 pKa = 11.84TPAVWCTIRR283 pKa = 11.84GIAMLIATDD292 pKa = 3.97SLLWWLEE299 pKa = 3.88CALVMAIAPQLGSTAPVVPQII320 pKa = 4.18
MM1 pKa = 7.82DD2 pKa = 5.28SDD4 pKa = 4.27MTTMHH9 pKa = 6.94HH10 pKa = 6.22SLSGSSQCPATLASPQLLARR30 pKa = 11.84SVDD33 pKa = 3.76AILRR37 pKa = 11.84PDD39 pKa = 3.39HH40 pKa = 6.58VFSLSRR46 pKa = 11.84LVSISLPGIYY56 pKa = 9.21TRR58 pKa = 11.84PHH60 pKa = 5.27TKK62 pKa = 9.96GVNGVGLGPWYY73 pKa = 10.32RR74 pKa = 11.84AVLGLLPPNYY84 pKa = 9.87EE85 pKa = 3.95NLLCRR90 pKa = 11.84RR91 pKa = 11.84LGVQSIWASPIDD103 pKa = 4.27LLQPPAASVRR113 pKa = 11.84VCSKK117 pKa = 10.15ISTSGSFPAADD128 pKa = 3.9FRR130 pKa = 11.84IALIGRR136 pKa = 11.84MPQQLFVHH144 pKa = 6.67LSSAPRR150 pKa = 11.84CHH152 pKa = 6.92DD153 pKa = 3.23VALRR157 pKa = 11.84RR158 pKa = 11.84SPKK161 pKa = 10.04LIMRR165 pKa = 11.84STHH168 pKa = 6.28PPGSFCAPNLQARR181 pKa = 11.84SMWASWAAGHH191 pKa = 5.95QRR193 pKa = 11.84VNHH196 pKa = 6.1GLPLATWGLGQRR208 pKa = 11.84LKK210 pKa = 10.01TLPGDD215 pKa = 3.11SRR217 pKa = 11.84YY218 pKa = 9.54RR219 pKa = 11.84VSRR222 pKa = 11.84AGFTGPHH229 pKa = 5.98KK230 pKa = 10.72CWLTGMAKK238 pKa = 10.12KK239 pKa = 9.91RR240 pKa = 11.84DD241 pKa = 3.88KK242 pKa = 9.93PTEE245 pKa = 3.68LAEE248 pKa = 4.05QGRR251 pKa = 11.84LQIPRR256 pKa = 11.84TLSDD260 pKa = 3.54LGGSSSQNPALIGRR274 pKa = 11.84TPAVWCTIRR283 pKa = 11.84GIAMLIATDD292 pKa = 3.97SLLWWLEE299 pKa = 3.88CALVMAIAPQLGSTAPVVPQII320 pKa = 4.18
Molecular weight: 34.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6232127 |
26 |
5754 |
474.4 |
52.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.218 ± 0.019 | 1.195 ± 0.008 |
5.886 ± 0.016 | 5.931 ± 0.02 |
3.628 ± 0.013 | 7.458 ± 0.02 |
2.379 ± 0.01 | 4.471 ± 0.015 |
4.581 ± 0.021 | 8.643 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.188 ± 0.007 | 3.553 ± 0.011 |
6.233 ± 0.023 | 4.028 ± 0.015 |
6.147 ± 0.019 | 8.102 ± 0.026 |
5.927 ± 0.015 | 6.271 ± 0.016 |
1.508 ± 0.008 | 2.649 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
