
Human plasma-associated gemycircularvirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemyvongvirus; Gemyvongvirus humas1; Human associated gemyvongvirus 1
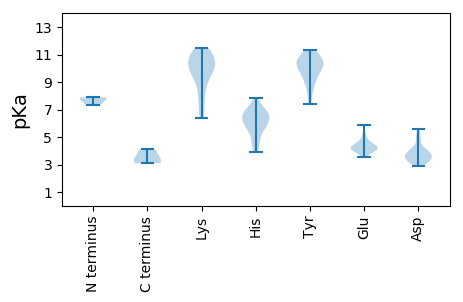
Average proteome isoelectric point is 7.36
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
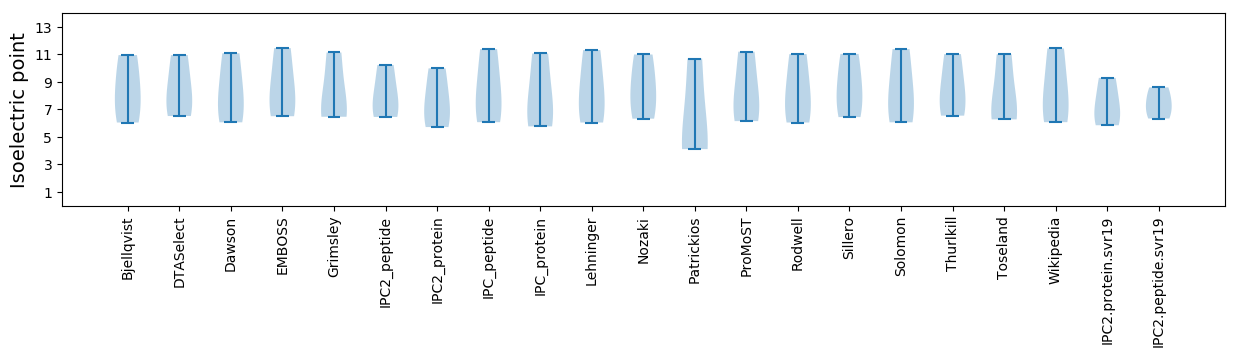
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0S2C4E4|A0A0S2C4E4_9VIRU Replication-associated protein OS=Human plasma-associated gemycircularvirus OX=1707330 GN=Rep PE=3 SV=1
MM1 pKa = 7.36TNVFPCWSSVTEE13 pKa = 4.19NPSEE17 pKa = 4.63AMTQFQEE24 pKa = 4.43LVSDD28 pKa = 4.25SPRR31 pKa = 11.84GFASSLTVRR40 pKa = 11.84HH41 pKa = 5.9RR42 pKa = 11.84QTLMLTQLCEE52 pKa = 4.37YY53 pKa = 8.53YY54 pKa = 10.41TEE56 pKa = 4.2TAEE59 pKa = 4.15VALSHH64 pKa = 6.87EE65 pKa = 4.39SHH67 pKa = 6.05TWMEE71 pKa = 4.09ALIIMLSWTTEE82 pKa = 3.76HH83 pKa = 7.03LGIGQTLVDD92 pKa = 4.91GMYY95 pKa = 10.39LACTRR100 pKa = 11.84TLSQSRR106 pKa = 11.84EE107 pKa = 3.78LRR109 pKa = 11.84LMHH112 pKa = 6.05MHH114 pKa = 6.05TSEE117 pKa = 3.96RR118 pKa = 11.84TRR120 pKa = 11.84TLFMSVLRR128 pKa = 11.84KK129 pKa = 9.52PLDD132 pKa = 3.15QGLEE136 pKa = 3.99DD137 pKa = 3.76RR138 pKa = 11.84LEE140 pKa = 4.02MLKK143 pKa = 10.6EE144 pKa = 3.52IANAHH149 pKa = 5.63GLRR152 pKa = 11.84SQMHH156 pKa = 3.92QTKK159 pKa = 9.73TSFLRR164 pKa = 11.84HH165 pKa = 4.43ARR167 pKa = 11.84RR168 pKa = 11.84WIRR171 pKa = 11.84EE172 pKa = 4.2VLSHH176 pKa = 7.31ASGTFPDD183 pKa = 3.94TQITPFQVYY192 pKa = 9.86EE193 pKa = 4.79DD194 pKa = 3.87NTSPQPGSPLDD205 pKa = 3.49SLSIHH210 pKa = 6.47DD211 pKa = 5.56LEE213 pKa = 5.49NISPIGSDD221 pKa = 3.25NEE223 pKa = 4.4GGKK226 pKa = 7.24TTLTLTLTRR235 pKa = 11.84KK236 pKa = 10.27GNEE239 pKa = 4.19LGPNGPGLSPNPLTLALTLTLTLANQLGQTYY270 pKa = 7.94LTHH273 pKa = 7.87AMGSFQNRR281 pKa = 11.84EE282 pKa = 4.0DD283 pKa = 3.37PTRR286 pKa = 11.84PLVRR290 pKa = 11.84TAA292 pKa = 4.1
MM1 pKa = 7.36TNVFPCWSSVTEE13 pKa = 4.19NPSEE17 pKa = 4.63AMTQFQEE24 pKa = 4.43LVSDD28 pKa = 4.25SPRR31 pKa = 11.84GFASSLTVRR40 pKa = 11.84HH41 pKa = 5.9RR42 pKa = 11.84QTLMLTQLCEE52 pKa = 4.37YY53 pKa = 8.53YY54 pKa = 10.41TEE56 pKa = 4.2TAEE59 pKa = 4.15VALSHH64 pKa = 6.87EE65 pKa = 4.39SHH67 pKa = 6.05TWMEE71 pKa = 4.09ALIIMLSWTTEE82 pKa = 3.76HH83 pKa = 7.03LGIGQTLVDD92 pKa = 4.91GMYY95 pKa = 10.39LACTRR100 pKa = 11.84TLSQSRR106 pKa = 11.84EE107 pKa = 3.78LRR109 pKa = 11.84LMHH112 pKa = 6.05MHH114 pKa = 6.05TSEE117 pKa = 3.96RR118 pKa = 11.84TRR120 pKa = 11.84TLFMSVLRR128 pKa = 11.84KK129 pKa = 9.52PLDD132 pKa = 3.15QGLEE136 pKa = 3.99DD137 pKa = 3.76RR138 pKa = 11.84LEE140 pKa = 4.02MLKK143 pKa = 10.6EE144 pKa = 3.52IANAHH149 pKa = 5.63GLRR152 pKa = 11.84SQMHH156 pKa = 3.92QTKK159 pKa = 9.73TSFLRR164 pKa = 11.84HH165 pKa = 4.43ARR167 pKa = 11.84RR168 pKa = 11.84WIRR171 pKa = 11.84EE172 pKa = 4.2VLSHH176 pKa = 7.31ASGTFPDD183 pKa = 3.94TQITPFQVYY192 pKa = 9.86EE193 pKa = 4.79DD194 pKa = 3.87NTSPQPGSPLDD205 pKa = 3.49SLSIHH210 pKa = 6.47DD211 pKa = 5.56LEE213 pKa = 5.49NISPIGSDD221 pKa = 3.25NEE223 pKa = 4.4GGKK226 pKa = 7.24TTLTLTLTRR235 pKa = 11.84KK236 pKa = 10.27GNEE239 pKa = 4.19LGPNGPGLSPNPLTLALTLTLTLANQLGQTYY270 pKa = 7.94LTHH273 pKa = 7.87AMGSFQNRR281 pKa = 11.84EE282 pKa = 4.0DD283 pKa = 3.37PTRR286 pKa = 11.84PLVRR290 pKa = 11.84TAA292 pKa = 4.1
Molecular weight: 32.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2C3Q0|A0A0S2C3Q0_9VIRU Putative ORF3 protein OS=Human plasma-associated gemycircularvirus OX=1707330 GN=ORF3 PE=4 SV=1
MM1 pKa = 7.8AYY3 pKa = 10.21ARR5 pKa = 11.84YY6 pKa = 9.31RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84PYY12 pKa = 8.53RR13 pKa = 11.84TNRR16 pKa = 11.84KK17 pKa = 6.35RR18 pKa = 11.84TSYY21 pKa = 10.41RR22 pKa = 11.84RR23 pKa = 11.84NTRR26 pKa = 11.84RR27 pKa = 11.84TRR29 pKa = 11.84NAPFRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SYY38 pKa = 10.43RR39 pKa = 11.84RR40 pKa = 11.84PANRR44 pKa = 11.84FKK46 pKa = 11.08KK47 pKa = 9.92RR48 pKa = 11.84VLAISSKK55 pKa = 10.93KK56 pKa = 10.59KK57 pKa = 9.76FDD59 pKa = 3.42TMRR62 pKa = 11.84SGSANGVIDD71 pKa = 3.45VTFNDD76 pKa = 3.41VQQAIMAWSPTTIPYY91 pKa = 10.53NEE93 pKa = 4.57VEE95 pKa = 4.44HH96 pKa = 7.04SPAHH100 pKa = 6.59RR101 pKa = 11.84NSQQIFFTGVKK112 pKa = 8.3EE113 pKa = 4.22TLDD116 pKa = 3.78PYY118 pKa = 11.21AIFPFHH124 pKa = 6.4WRR126 pKa = 11.84RR127 pKa = 11.84VVFWTHH133 pKa = 4.45EE134 pKa = 3.97RR135 pKa = 11.84FPQFEE140 pKa = 4.61TIVAAGGTRR149 pKa = 11.84FRR151 pKa = 11.84YY152 pKa = 9.2GVTYY156 pKa = 10.42GVTDD160 pKa = 3.69TPVQAIQEE168 pKa = 4.21LMFQGTIGIDD178 pKa = 3.37WDD180 pKa = 3.55PTRR183 pKa = 11.84PMDD186 pKa = 4.79ARR188 pKa = 11.84LDD190 pKa = 3.42RR191 pKa = 11.84HH192 pKa = 6.38RR193 pKa = 11.84IKK195 pKa = 10.44IVSDD199 pKa = 2.89KK200 pKa = 10.71RR201 pKa = 11.84VNFNPQSDD209 pKa = 3.45NGRR212 pKa = 11.84YY213 pKa = 9.15SVFKK217 pKa = 8.88RR218 pKa = 11.84WHH220 pKa = 5.55SIRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84IMYY228 pKa = 9.44DD229 pKa = 3.05QEE231 pKa = 4.12EE232 pKa = 4.47AGDD235 pKa = 4.04DD236 pKa = 3.79QISDD240 pKa = 3.33VWSTVAPTSSGNLFIVDD257 pKa = 4.0FFGFPPGLAQGAAARR272 pKa = 11.84LGLQSTVYY280 pKa = 9.57WRR282 pKa = 11.84EE283 pKa = 3.74SS284 pKa = 3.1
MM1 pKa = 7.8AYY3 pKa = 10.21ARR5 pKa = 11.84YY6 pKa = 9.31RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84PYY12 pKa = 8.53RR13 pKa = 11.84TNRR16 pKa = 11.84KK17 pKa = 6.35RR18 pKa = 11.84TSYY21 pKa = 10.41RR22 pKa = 11.84RR23 pKa = 11.84NTRR26 pKa = 11.84RR27 pKa = 11.84TRR29 pKa = 11.84NAPFRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84SYY38 pKa = 10.43RR39 pKa = 11.84RR40 pKa = 11.84PANRR44 pKa = 11.84FKK46 pKa = 11.08KK47 pKa = 9.92RR48 pKa = 11.84VLAISSKK55 pKa = 10.93KK56 pKa = 10.59KK57 pKa = 9.76FDD59 pKa = 3.42TMRR62 pKa = 11.84SGSANGVIDD71 pKa = 3.45VTFNDD76 pKa = 3.41VQQAIMAWSPTTIPYY91 pKa = 10.53NEE93 pKa = 4.57VEE95 pKa = 4.44HH96 pKa = 7.04SPAHH100 pKa = 6.59RR101 pKa = 11.84NSQQIFFTGVKK112 pKa = 8.3EE113 pKa = 4.22TLDD116 pKa = 3.78PYY118 pKa = 11.21AIFPFHH124 pKa = 6.4WRR126 pKa = 11.84RR127 pKa = 11.84VVFWTHH133 pKa = 4.45EE134 pKa = 3.97RR135 pKa = 11.84FPQFEE140 pKa = 4.61TIVAAGGTRR149 pKa = 11.84FRR151 pKa = 11.84YY152 pKa = 9.2GVTYY156 pKa = 10.42GVTDD160 pKa = 3.69TPVQAIQEE168 pKa = 4.21LMFQGTIGIDD178 pKa = 3.37WDD180 pKa = 3.55PTRR183 pKa = 11.84PMDD186 pKa = 4.79ARR188 pKa = 11.84LDD190 pKa = 3.42RR191 pKa = 11.84HH192 pKa = 6.38RR193 pKa = 11.84IKK195 pKa = 10.44IVSDD199 pKa = 2.89KK200 pKa = 10.71RR201 pKa = 11.84VNFNPQSDD209 pKa = 3.45NGRR212 pKa = 11.84YY213 pKa = 9.15SVFKK217 pKa = 8.88RR218 pKa = 11.84WHH220 pKa = 5.55SIRR223 pKa = 11.84RR224 pKa = 11.84RR225 pKa = 11.84IMYY228 pKa = 9.44DD229 pKa = 3.05QEE231 pKa = 4.12EE232 pKa = 4.47AGDD235 pKa = 4.04DD236 pKa = 3.79QISDD240 pKa = 3.33VWSTVAPTSSGNLFIVDD257 pKa = 4.0FFGFPPGLAQGAAARR272 pKa = 11.84LGLQSTVYY280 pKa = 9.57WRR282 pKa = 11.84EE283 pKa = 3.74SS284 pKa = 3.1
Molecular weight: 33.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
917 |
284 |
341 |
305.7 |
35.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.78 ± 0.471 | 1.2 ± 0.617 |
5.78 ± 1.233 | 4.689 ± 0.842 |
5.016 ± 0.889 | 6.979 ± 1.094 |
2.944 ± 0.455 | 4.253 ± 0.529 |
2.835 ± 0.44 | 7.197 ± 2.535 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.29 ± 0.868 | 3.926 ± 0.108 |
5.562 ± 0.033 | 4.253 ± 0.554 |
9.815 ± 1.498 | 7.415 ± 0.597 |
8.724 ± 1.504 | 5.453 ± 0.794 |
2.181 ± 0.341 | 3.708 ± 0.874 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
