
Sclerotinia sclerotiorum negative-stranded RNA virus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Mymonaviridae; Sclerotimonavirus; Drop sclerotimonavirus
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
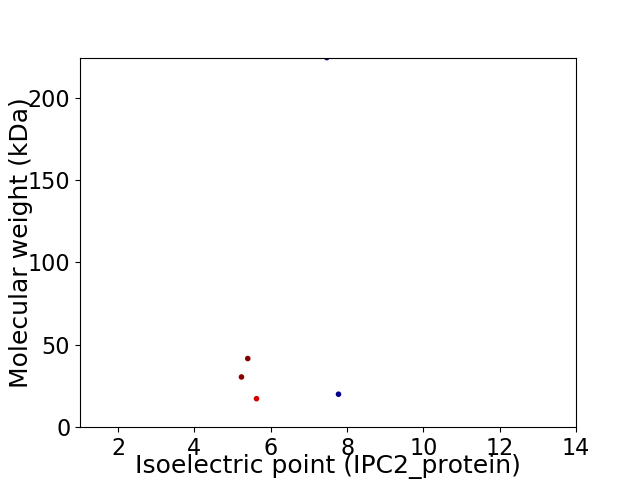
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M4LD35|A0A0M4LD35_9MONO Gp1 OS=Sclerotinia sclerotiorum negative-stranded RNA virus 4 OX=1708391 PE=4 SV=1
MM1 pKa = 7.78ASKK4 pKa = 10.52QNPQVISEE12 pKa = 4.23EE13 pKa = 4.2AVASLDD19 pKa = 3.67NNVSGRR25 pKa = 11.84SFSMGVSKK33 pKa = 10.49MMSSLSRR40 pKa = 11.84ASWADD45 pKa = 3.33SVEE48 pKa = 4.08EE49 pKa = 4.44EE50 pKa = 4.81LPKK53 pKa = 11.04SPDD56 pKa = 3.05STAKK60 pKa = 9.42LTTIGSAKK68 pKa = 10.17LASTSPARR76 pKa = 11.84IDD78 pKa = 3.41SQSRR82 pKa = 11.84SDD84 pKa = 3.57APEE87 pKa = 4.12SEE89 pKa = 4.73SEE91 pKa = 5.01DD92 pKa = 3.56EE93 pKa = 4.43FQEE96 pKa = 4.32SPVIKK101 pKa = 10.65KK102 pKa = 10.05EE103 pKa = 3.94GSQLAAEE110 pKa = 4.18EE111 pKa = 4.79SEE113 pKa = 5.14VEE115 pKa = 4.38FNPGPVVKK123 pKa = 10.69GSAPKK128 pKa = 10.0QAVGILGVRR137 pKa = 11.84GNQRR141 pKa = 11.84EE142 pKa = 4.23ILNPTAQKK150 pKa = 8.98ITDD153 pKa = 3.69EE154 pKa = 4.14QLAGTTMKK162 pKa = 9.25ITLDD166 pKa = 3.22KK167 pKa = 10.44LQQVVEE173 pKa = 4.24AQDD176 pKa = 3.57NVIKK180 pKa = 10.7ILLQRR185 pKa = 11.84LDD187 pKa = 3.65ASDD190 pKa = 3.55TKK192 pKa = 10.61IANLLATINSLKK204 pKa = 10.49VVAEE208 pKa = 4.31DD209 pKa = 3.23VSSRR213 pKa = 11.84VKK215 pKa = 10.92SIDD218 pKa = 3.41ADD220 pKa = 3.7ALDD223 pKa = 4.65VLNQAAKK230 pKa = 9.67IVRR233 pKa = 11.84DD234 pKa = 3.58NRR236 pKa = 11.84VPDD239 pKa = 3.61APIAKK244 pKa = 9.25IEE246 pKa = 4.02QEE248 pKa = 4.12LEE250 pKa = 3.88RR251 pKa = 11.84TQDD254 pKa = 3.46SVAKK258 pKa = 9.57AAEE261 pKa = 4.1RR262 pKa = 11.84APIQSPKK269 pKa = 10.11IKK271 pKa = 9.95ASKK274 pKa = 9.03GARR277 pKa = 11.84RR278 pKa = 11.84LKK280 pKa = 10.87LSFEE284 pKa = 4.37
MM1 pKa = 7.78ASKK4 pKa = 10.52QNPQVISEE12 pKa = 4.23EE13 pKa = 4.2AVASLDD19 pKa = 3.67NNVSGRR25 pKa = 11.84SFSMGVSKK33 pKa = 10.49MMSSLSRR40 pKa = 11.84ASWADD45 pKa = 3.33SVEE48 pKa = 4.08EE49 pKa = 4.44EE50 pKa = 4.81LPKK53 pKa = 11.04SPDD56 pKa = 3.05STAKK60 pKa = 9.42LTTIGSAKK68 pKa = 10.17LASTSPARR76 pKa = 11.84IDD78 pKa = 3.41SQSRR82 pKa = 11.84SDD84 pKa = 3.57APEE87 pKa = 4.12SEE89 pKa = 4.73SEE91 pKa = 5.01DD92 pKa = 3.56EE93 pKa = 4.43FQEE96 pKa = 4.32SPVIKK101 pKa = 10.65KK102 pKa = 10.05EE103 pKa = 3.94GSQLAAEE110 pKa = 4.18EE111 pKa = 4.79SEE113 pKa = 5.14VEE115 pKa = 4.38FNPGPVVKK123 pKa = 10.69GSAPKK128 pKa = 10.0QAVGILGVRR137 pKa = 11.84GNQRR141 pKa = 11.84EE142 pKa = 4.23ILNPTAQKK150 pKa = 8.98ITDD153 pKa = 3.69EE154 pKa = 4.14QLAGTTMKK162 pKa = 9.25ITLDD166 pKa = 3.22KK167 pKa = 10.44LQQVVEE173 pKa = 4.24AQDD176 pKa = 3.57NVIKK180 pKa = 10.7ILLQRR185 pKa = 11.84LDD187 pKa = 3.65ASDD190 pKa = 3.55TKK192 pKa = 10.61IANLLATINSLKK204 pKa = 10.49VVAEE208 pKa = 4.31DD209 pKa = 3.23VSSRR213 pKa = 11.84VKK215 pKa = 10.92SIDD218 pKa = 3.41ADD220 pKa = 3.7ALDD223 pKa = 4.65VLNQAAKK230 pKa = 9.67IVRR233 pKa = 11.84DD234 pKa = 3.58NRR236 pKa = 11.84VPDD239 pKa = 3.61APIAKK244 pKa = 9.25IEE246 pKa = 4.02QEE248 pKa = 4.12LEE250 pKa = 3.88RR251 pKa = 11.84TQDD254 pKa = 3.46SVAKK258 pKa = 9.57AAEE261 pKa = 4.1RR262 pKa = 11.84APIQSPKK269 pKa = 10.11IKK271 pKa = 9.95ASKK274 pKa = 9.03GARR277 pKa = 11.84RR278 pKa = 11.84LKK280 pKa = 10.87LSFEE284 pKa = 4.37
Molecular weight: 30.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M4KRD1|A0A0M4KRD1_9MONO Gp2 OS=Sclerotinia sclerotiorum negative-stranded RNA virus 4 OX=1708391 PE=4 SV=1
MM1 pKa = 7.37SRR3 pKa = 11.84LRR5 pKa = 11.84LPQSFSGPVWTFSILGVIVSKK26 pKa = 10.78RR27 pKa = 11.84PEE29 pKa = 3.76ILLQTRR35 pKa = 11.84LAALRR40 pKa = 11.84TLILVSRR47 pKa = 11.84STSLSTRR54 pKa = 11.84VSMLRR59 pKa = 11.84LPIEE63 pKa = 4.4VIAPFSPEE71 pKa = 3.23MTAVHH76 pKa = 6.52EE77 pKa = 4.72FCEE80 pKa = 3.95QLPDD84 pKa = 4.61HH85 pKa = 6.93ISVSDD90 pKa = 3.85LNRR93 pKa = 11.84VIDD96 pKa = 3.66IGLAGLTVAPSTLATYY112 pKa = 10.21IAVASVSEE120 pKa = 4.19HH121 pKa = 6.49LSCEE125 pKa = 4.12AKK127 pKa = 10.73VLTASLGIGEE137 pKa = 4.83LPRR140 pKa = 11.84SRR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84EE145 pKa = 3.4IRR147 pKa = 11.84FEE149 pKa = 3.88VVRR152 pKa = 11.84ILNEE156 pKa = 3.78SDD158 pKa = 3.03VTLRR162 pKa = 11.84DD163 pKa = 3.51KK164 pKa = 11.47VKK166 pKa = 10.89LIQQDD171 pKa = 3.47ILSSSIPSATEE182 pKa = 3.37
MM1 pKa = 7.37SRR3 pKa = 11.84LRR5 pKa = 11.84LPQSFSGPVWTFSILGVIVSKK26 pKa = 10.78RR27 pKa = 11.84PEE29 pKa = 3.76ILLQTRR35 pKa = 11.84LAALRR40 pKa = 11.84TLILVSRR47 pKa = 11.84STSLSTRR54 pKa = 11.84VSMLRR59 pKa = 11.84LPIEE63 pKa = 4.4VIAPFSPEE71 pKa = 3.23MTAVHH76 pKa = 6.52EE77 pKa = 4.72FCEE80 pKa = 3.95QLPDD84 pKa = 4.61HH85 pKa = 6.93ISVSDD90 pKa = 3.85LNRR93 pKa = 11.84VIDD96 pKa = 3.66IGLAGLTVAPSTLATYY112 pKa = 10.21IAVASVSEE120 pKa = 4.19HH121 pKa = 6.49LSCEE125 pKa = 4.12AKK127 pKa = 10.73VLTASLGIGEE137 pKa = 4.83LPRR140 pKa = 11.84SRR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84EE145 pKa = 3.4IRR147 pKa = 11.84FEE149 pKa = 3.88VVRR152 pKa = 11.84ILNEE156 pKa = 3.78SDD158 pKa = 3.03VTLRR162 pKa = 11.84DD163 pKa = 3.51KK164 pKa = 11.47VKK166 pKa = 10.89LIQQDD171 pKa = 3.47ILSSSIPSATEE182 pKa = 3.37
Molecular weight: 20.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3015 |
158 |
2010 |
603.0 |
66.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.894 ± 1.319 | 1.194 ± 0.436 |
5.373 ± 0.468 | 5.904 ± 0.842 |
3.549 ± 0.537 | 5.771 ± 0.719 |
1.725 ± 0.4 | 6.103 ± 0.516 |
4.71 ± 0.794 | 10.912 ± 0.72 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.454 ± 0.145 | 3.018 ± 0.348 |
5.274 ± 0.412 | 4.046 ± 0.43 |
6.534 ± 0.648 | 9.983 ± 0.74 |
5.274 ± 0.409 | 6.866 ± 0.36 |
1.327 ± 0.382 | 2.09 ± 0.576 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
