
Capybara microvirus Cap3_SP_441
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
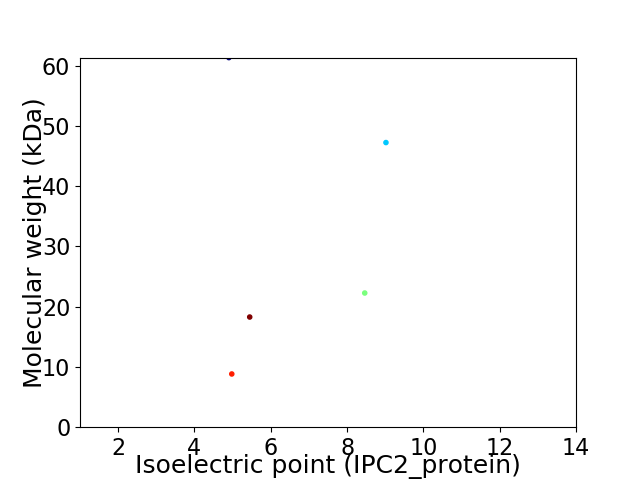
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5X1|A0A4P8W5X1_9VIRU Nonstructural protein OS=Capybara microvirus Cap3_SP_441 OX=2585453 PE=4 SV=1
MM1 pKa = 7.35SRR3 pKa = 11.84NEE5 pKa = 3.64LAYY8 pKa = 10.03EE9 pKa = 4.04VLDD12 pKa = 4.63TIDD15 pKa = 3.87TPRR18 pKa = 11.84STWQRR23 pKa = 11.84NFTHH27 pKa = 5.98KK28 pKa = 9.74TSFNAGKK35 pKa = 10.27LIPFYY40 pKa = 10.25IDD42 pKa = 2.91TDD44 pKa = 4.38IIPGTTIKK52 pKa = 10.78NKK54 pKa = 7.91TSIVIRR60 pKa = 11.84MSTPIAPVMDD70 pKa = 4.2NLYY73 pKa = 10.83LDD75 pKa = 4.06TYY77 pKa = 10.16YY78 pKa = 10.85FKK80 pKa = 10.92CSKK83 pKa = 9.39FWYY86 pKa = 8.05WEE88 pKa = 3.68HH89 pKa = 6.45FRR91 pKa = 11.84AQMGEE96 pKa = 4.08NNQSAWTQTIEE107 pKa = 4.47YY108 pKa = 9.45IEE110 pKa = 4.36PQIKK114 pKa = 8.49MLSMEE119 pKa = 4.1YY120 pKa = 10.41GANDD124 pKa = 3.34VACYY128 pKa = 9.56MGMPINVANLEE139 pKa = 4.18WSKK142 pKa = 11.35LPVQAYY148 pKa = 9.67IDD150 pKa = 3.21IWNNWFRR157 pKa = 11.84DD158 pKa = 3.68EE159 pKa = 4.63NLQAPITLDD168 pKa = 3.21KK169 pKa = 11.01TDD171 pKa = 4.44ADD173 pKa = 4.01LTADD177 pKa = 3.42ATINTGFGLLPVCKK191 pKa = 9.86FHH193 pKa = 8.39DD194 pKa = 4.49YY195 pKa = 8.25FTSLLPQPQKK205 pKa = 11.38GEE207 pKa = 4.09AVTMPLGVAAPVSGTVKK224 pKa = 10.85GNGLQLGVTNGTTNFVPTTRR244 pKa = 11.84GSNYY248 pKa = 10.22ASNTFGDD255 pKa = 4.02SLGSFSEE262 pKa = 4.34FTPGGTKK269 pKa = 9.9TYY271 pKa = 10.66GVTPDD276 pKa = 3.36EE277 pKa = 4.69SKK279 pKa = 10.89SGLVIGTAYY288 pKa = 10.71ADD290 pKa = 3.79LTSATAATFNALRR303 pKa = 11.84LAGATQRR310 pKa = 11.84ILEE313 pKa = 4.15KK314 pKa = 10.39DD315 pKa = 3.0ARR317 pKa = 11.84FGTRR321 pKa = 11.84YY322 pKa = 10.0PEE324 pKa = 4.04FLKK327 pKa = 10.96GQYY330 pKa = 10.34GVTAADD336 pKa = 3.67EE337 pKa = 4.4CLLIPEE343 pKa = 4.23YY344 pKa = 10.85LGGKK348 pKa = 9.42RR349 pKa = 11.84IPINIEE355 pKa = 3.84QVCQTSSTDD364 pKa = 3.15STSPLGEE371 pKa = 4.06TGAMSVTADD380 pKa = 3.13INEE383 pKa = 4.73DD384 pKa = 3.43FTKK387 pKa = 10.84SFTKK391 pKa = 10.11HH392 pKa = 6.27DD393 pKa = 3.95FLIGVCCVRR402 pKa = 11.84AEE404 pKa = 3.94HH405 pKa = 6.96TYY407 pKa = 9.94QQGIPLQFKK416 pKa = 9.92RR417 pKa = 11.84KK418 pKa = 9.03RR419 pKa = 11.84RR420 pKa = 11.84LDD422 pKa = 3.49TYY424 pKa = 10.51HH425 pKa = 7.55PSLAHH430 pKa = 6.92IGNQPVYY437 pKa = 10.03NFEE440 pKa = 4.34IFAQGNSTDD449 pKa = 4.19DD450 pKa = 3.11EE451 pKa = 4.97VMGYY455 pKa = 9.92KK456 pKa = 9.81EE457 pKa = 3.85AWQEE461 pKa = 3.68YY462 pKa = 9.17LYY464 pKa = 10.9KK465 pKa = 10.26PNRR468 pKa = 11.84ISGQMLSTYY477 pKa = 7.29TQSLDD482 pKa = 2.93VWHH485 pKa = 6.74YY486 pKa = 10.96GDD488 pKa = 5.8DD489 pKa = 4.11YY490 pKa = 12.05VNLPVLSSEE499 pKa = 4.47WIVEE503 pKa = 3.72PTEE506 pKa = 4.26YY507 pKa = 9.51IDD509 pKa = 3.24RR510 pKa = 11.84TLAVSSSVSNQFWADD525 pKa = 2.99IYY527 pKa = 10.83IEE529 pKa = 4.41QIVSAPIPINRR540 pKa = 11.84VPGLIDD546 pKa = 3.56HH547 pKa = 6.51YY548 pKa = 11.72
MM1 pKa = 7.35SRR3 pKa = 11.84NEE5 pKa = 3.64LAYY8 pKa = 10.03EE9 pKa = 4.04VLDD12 pKa = 4.63TIDD15 pKa = 3.87TPRR18 pKa = 11.84STWQRR23 pKa = 11.84NFTHH27 pKa = 5.98KK28 pKa = 9.74TSFNAGKK35 pKa = 10.27LIPFYY40 pKa = 10.25IDD42 pKa = 2.91TDD44 pKa = 4.38IIPGTTIKK52 pKa = 10.78NKK54 pKa = 7.91TSIVIRR60 pKa = 11.84MSTPIAPVMDD70 pKa = 4.2NLYY73 pKa = 10.83LDD75 pKa = 4.06TYY77 pKa = 10.16YY78 pKa = 10.85FKK80 pKa = 10.92CSKK83 pKa = 9.39FWYY86 pKa = 8.05WEE88 pKa = 3.68HH89 pKa = 6.45FRR91 pKa = 11.84AQMGEE96 pKa = 4.08NNQSAWTQTIEE107 pKa = 4.47YY108 pKa = 9.45IEE110 pKa = 4.36PQIKK114 pKa = 8.49MLSMEE119 pKa = 4.1YY120 pKa = 10.41GANDD124 pKa = 3.34VACYY128 pKa = 9.56MGMPINVANLEE139 pKa = 4.18WSKK142 pKa = 11.35LPVQAYY148 pKa = 9.67IDD150 pKa = 3.21IWNNWFRR157 pKa = 11.84DD158 pKa = 3.68EE159 pKa = 4.63NLQAPITLDD168 pKa = 3.21KK169 pKa = 11.01TDD171 pKa = 4.44ADD173 pKa = 4.01LTADD177 pKa = 3.42ATINTGFGLLPVCKK191 pKa = 9.86FHH193 pKa = 8.39DD194 pKa = 4.49YY195 pKa = 8.25FTSLLPQPQKK205 pKa = 11.38GEE207 pKa = 4.09AVTMPLGVAAPVSGTVKK224 pKa = 10.85GNGLQLGVTNGTTNFVPTTRR244 pKa = 11.84GSNYY248 pKa = 10.22ASNTFGDD255 pKa = 4.02SLGSFSEE262 pKa = 4.34FTPGGTKK269 pKa = 9.9TYY271 pKa = 10.66GVTPDD276 pKa = 3.36EE277 pKa = 4.69SKK279 pKa = 10.89SGLVIGTAYY288 pKa = 10.71ADD290 pKa = 3.79LTSATAATFNALRR303 pKa = 11.84LAGATQRR310 pKa = 11.84ILEE313 pKa = 4.15KK314 pKa = 10.39DD315 pKa = 3.0ARR317 pKa = 11.84FGTRR321 pKa = 11.84YY322 pKa = 10.0PEE324 pKa = 4.04FLKK327 pKa = 10.96GQYY330 pKa = 10.34GVTAADD336 pKa = 3.67EE337 pKa = 4.4CLLIPEE343 pKa = 4.23YY344 pKa = 10.85LGGKK348 pKa = 9.42RR349 pKa = 11.84IPINIEE355 pKa = 3.84QVCQTSSTDD364 pKa = 3.15STSPLGEE371 pKa = 4.06TGAMSVTADD380 pKa = 3.13INEE383 pKa = 4.73DD384 pKa = 3.43FTKK387 pKa = 10.84SFTKK391 pKa = 10.11HH392 pKa = 6.27DD393 pKa = 3.95FLIGVCCVRR402 pKa = 11.84AEE404 pKa = 3.94HH405 pKa = 6.96TYY407 pKa = 9.94QQGIPLQFKK416 pKa = 9.92RR417 pKa = 11.84KK418 pKa = 9.03RR419 pKa = 11.84RR420 pKa = 11.84LDD422 pKa = 3.49TYY424 pKa = 10.51HH425 pKa = 7.55PSLAHH430 pKa = 6.92IGNQPVYY437 pKa = 10.03NFEE440 pKa = 4.34IFAQGNSTDD449 pKa = 4.19DD450 pKa = 3.11EE451 pKa = 4.97VMGYY455 pKa = 9.92KK456 pKa = 9.81EE457 pKa = 3.85AWQEE461 pKa = 3.68YY462 pKa = 9.17LYY464 pKa = 10.9KK465 pKa = 10.26PNRR468 pKa = 11.84ISGQMLSTYY477 pKa = 7.29TQSLDD482 pKa = 2.93VWHH485 pKa = 6.74YY486 pKa = 10.96GDD488 pKa = 5.8DD489 pKa = 4.11YY490 pKa = 12.05VNLPVLSSEE499 pKa = 4.47WIVEE503 pKa = 3.72PTEE506 pKa = 4.26YY507 pKa = 9.51IDD509 pKa = 3.24RR510 pKa = 11.84TLAVSSSVSNQFWADD525 pKa = 2.99IYY527 pKa = 10.83IEE529 pKa = 4.41QIVSAPIPINRR540 pKa = 11.84VPGLIDD546 pKa = 3.56HH547 pKa = 6.51YY548 pKa = 11.72
Molecular weight: 61.34 kDa
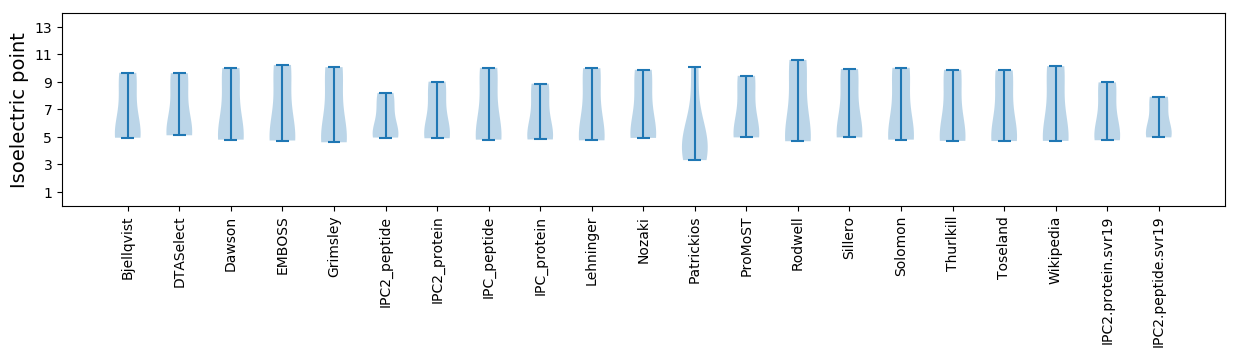
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W8K7|A0A4P8W8K7_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_441 OX=2585453 PE=4 SV=1
MM1 pKa = 7.81HH2 pKa = 6.78YY3 pKa = 10.63NCRR6 pKa = 11.84KK7 pKa = 10.19NKK9 pKa = 9.67GLQEE13 pKa = 4.04QNSIMLSVGTNKK25 pKa = 10.22SSGRR29 pKa = 11.84VPTFFIKK36 pKa = 10.46EE37 pKa = 4.06SRR39 pKa = 11.84EE40 pKa = 4.09MVCLNPITAWKK51 pKa = 10.23KK52 pKa = 10.41KK53 pKa = 10.11FITLGTNPQFYY64 pKa = 9.92EE65 pKa = 4.09YY66 pKa = 10.3EE67 pKa = 3.91KK68 pKa = 10.7KK69 pKa = 10.28IRR71 pKa = 11.84FKK73 pKa = 11.33NPNDD77 pKa = 3.38EE78 pKa = 4.15NFEE81 pKa = 4.24QIQLPCGKK89 pKa = 10.1CLGCRR94 pKa = 11.84LDD96 pKa = 5.06HH97 pKa = 6.61ANAWATRR104 pKa = 11.84IMLEE108 pKa = 4.43AKK110 pKa = 9.95NHH112 pKa = 5.5EE113 pKa = 4.52KK114 pKa = 10.79NCFITLTYY122 pKa = 10.3NPKK125 pKa = 10.11NLPLTEE131 pKa = 4.15TGKK134 pKa = 8.13MTLKK138 pKa = 10.7KK139 pKa = 10.44SDD141 pKa = 2.96IQKK144 pKa = 9.2FFKK147 pKa = 10.53RR148 pKa = 11.84LRR150 pKa = 11.84EE151 pKa = 3.95KK152 pKa = 10.34TKK154 pKa = 10.84LKK156 pKa = 10.71LSYY159 pKa = 9.68FCCGEE164 pKa = 3.78YY165 pKa = 10.73GPRR168 pKa = 11.84TFRR171 pKa = 11.84PHH173 pKa = 3.87YY174 pKa = 8.84HH175 pKa = 6.92AIIFGYY181 pKa = 10.14EE182 pKa = 3.78PSDD185 pKa = 3.33LKK187 pKa = 11.24LRR189 pKa = 11.84AFSKK193 pKa = 9.71TNQKK197 pKa = 9.07MFEE200 pKa = 4.15SEE202 pKa = 4.36EE203 pKa = 3.97LLKK206 pKa = 10.09TWGLGFITIEE216 pKa = 3.75EE217 pKa = 4.53LNYY220 pKa = 9.19KK221 pKa = 6.61TACYY225 pKa = 7.13TARR228 pKa = 11.84YY229 pKa = 6.98VQKK232 pKa = 10.82KK233 pKa = 10.37AGLKK237 pKa = 8.07TNSRR241 pKa = 11.84KK242 pKa = 8.33FTGATEE248 pKa = 4.21TKK250 pKa = 10.14IKK252 pKa = 9.98IDD254 pKa = 3.38EE255 pKa = 4.35RR256 pKa = 11.84TGDD259 pKa = 3.79EE260 pKa = 4.19YY261 pKa = 11.63LSTINKK267 pKa = 9.75LKK269 pKa = 10.08TEE271 pKa = 3.95KK272 pKa = 10.17FDD274 pKa = 3.3NYY276 pKa = 10.37GRR278 pKa = 11.84EE279 pKa = 3.87KK280 pKa = 10.93EE281 pKa = 4.29FIVMSKK287 pKa = 10.44KK288 pKa = 9.99PAIGLNYY295 pKa = 9.38WLEE298 pKa = 4.15NKK300 pKa = 9.98EE301 pKa = 3.95KK302 pKa = 10.32ILRR305 pKa = 11.84NKK307 pKa = 10.19GILIKK312 pKa = 9.9IDD314 pKa = 3.45EE315 pKa = 4.53TVKK318 pKa = 10.68LKK320 pKa = 10.27PIPRR324 pKa = 11.84YY325 pKa = 8.98FEE327 pKa = 5.41KK328 pKa = 10.44IMEE331 pKa = 4.45RR332 pKa = 11.84EE333 pKa = 3.98NKK335 pKa = 7.57EE336 pKa = 3.82TLDD339 pKa = 3.65RR340 pKa = 11.84LKK342 pKa = 10.87HH343 pKa = 4.97EE344 pKa = 4.57KK345 pKa = 10.36KK346 pKa = 10.67EE347 pKa = 3.97NFIKK351 pKa = 10.67NQEE354 pKa = 4.37LIVKK358 pKa = 9.85KK359 pKa = 10.32IKK361 pKa = 10.94NNFKK365 pKa = 10.66LCEE368 pKa = 4.0EE369 pKa = 4.3LTNDD373 pKa = 4.32EE374 pKa = 4.09KK375 pKa = 10.91TKK377 pKa = 10.38IYY379 pKa = 11.14NNYY382 pKa = 9.54LNKK385 pKa = 10.46NLTNRR390 pKa = 11.84AKK392 pKa = 10.11YY393 pKa = 10.2LKK395 pKa = 10.17RR396 pKa = 11.84GQII399 pKa = 3.58
MM1 pKa = 7.81HH2 pKa = 6.78YY3 pKa = 10.63NCRR6 pKa = 11.84KK7 pKa = 10.19NKK9 pKa = 9.67GLQEE13 pKa = 4.04QNSIMLSVGTNKK25 pKa = 10.22SSGRR29 pKa = 11.84VPTFFIKK36 pKa = 10.46EE37 pKa = 4.06SRR39 pKa = 11.84EE40 pKa = 4.09MVCLNPITAWKK51 pKa = 10.23KK52 pKa = 10.41KK53 pKa = 10.11FITLGTNPQFYY64 pKa = 9.92EE65 pKa = 4.09YY66 pKa = 10.3EE67 pKa = 3.91KK68 pKa = 10.7KK69 pKa = 10.28IRR71 pKa = 11.84FKK73 pKa = 11.33NPNDD77 pKa = 3.38EE78 pKa = 4.15NFEE81 pKa = 4.24QIQLPCGKK89 pKa = 10.1CLGCRR94 pKa = 11.84LDD96 pKa = 5.06HH97 pKa = 6.61ANAWATRR104 pKa = 11.84IMLEE108 pKa = 4.43AKK110 pKa = 9.95NHH112 pKa = 5.5EE113 pKa = 4.52KK114 pKa = 10.79NCFITLTYY122 pKa = 10.3NPKK125 pKa = 10.11NLPLTEE131 pKa = 4.15TGKK134 pKa = 8.13MTLKK138 pKa = 10.7KK139 pKa = 10.44SDD141 pKa = 2.96IQKK144 pKa = 9.2FFKK147 pKa = 10.53RR148 pKa = 11.84LRR150 pKa = 11.84EE151 pKa = 3.95KK152 pKa = 10.34TKK154 pKa = 10.84LKK156 pKa = 10.71LSYY159 pKa = 9.68FCCGEE164 pKa = 3.78YY165 pKa = 10.73GPRR168 pKa = 11.84TFRR171 pKa = 11.84PHH173 pKa = 3.87YY174 pKa = 8.84HH175 pKa = 6.92AIIFGYY181 pKa = 10.14EE182 pKa = 3.78PSDD185 pKa = 3.33LKK187 pKa = 11.24LRR189 pKa = 11.84AFSKK193 pKa = 9.71TNQKK197 pKa = 9.07MFEE200 pKa = 4.15SEE202 pKa = 4.36EE203 pKa = 3.97LLKK206 pKa = 10.09TWGLGFITIEE216 pKa = 3.75EE217 pKa = 4.53LNYY220 pKa = 9.19KK221 pKa = 6.61TACYY225 pKa = 7.13TARR228 pKa = 11.84YY229 pKa = 6.98VQKK232 pKa = 10.82KK233 pKa = 10.37AGLKK237 pKa = 8.07TNSRR241 pKa = 11.84KK242 pKa = 8.33FTGATEE248 pKa = 4.21TKK250 pKa = 10.14IKK252 pKa = 9.98IDD254 pKa = 3.38EE255 pKa = 4.35RR256 pKa = 11.84TGDD259 pKa = 3.79EE260 pKa = 4.19YY261 pKa = 11.63LSTINKK267 pKa = 9.75LKK269 pKa = 10.08TEE271 pKa = 3.95KK272 pKa = 10.17FDD274 pKa = 3.3NYY276 pKa = 10.37GRR278 pKa = 11.84EE279 pKa = 3.87KK280 pKa = 10.93EE281 pKa = 4.29FIVMSKK287 pKa = 10.44KK288 pKa = 9.99PAIGLNYY295 pKa = 9.38WLEE298 pKa = 4.15NKK300 pKa = 9.98EE301 pKa = 3.95KK302 pKa = 10.32ILRR305 pKa = 11.84NKK307 pKa = 10.19GILIKK312 pKa = 9.9IDD314 pKa = 3.45EE315 pKa = 4.53TVKK318 pKa = 10.68LKK320 pKa = 10.27PIPRR324 pKa = 11.84YY325 pKa = 8.98FEE327 pKa = 5.41KK328 pKa = 10.44IMEE331 pKa = 4.45RR332 pKa = 11.84EE333 pKa = 3.98NKK335 pKa = 7.57EE336 pKa = 3.82TLDD339 pKa = 3.65RR340 pKa = 11.84LKK342 pKa = 10.87HH343 pKa = 4.97EE344 pKa = 4.57KK345 pKa = 10.36KK346 pKa = 10.67EE347 pKa = 3.97NFIKK351 pKa = 10.67NQEE354 pKa = 4.37LIVKK358 pKa = 9.85KK359 pKa = 10.32IKK361 pKa = 10.94NNFKK365 pKa = 10.66LCEE368 pKa = 4.0EE369 pKa = 4.3LTNDD373 pKa = 4.32EE374 pKa = 4.09KK375 pKa = 10.91TKK377 pKa = 10.38IYY379 pKa = 11.14NNYY382 pKa = 9.54LNKK385 pKa = 10.46NLTNRR390 pKa = 11.84AKK392 pKa = 10.11YY393 pKa = 10.2LKK395 pKa = 10.17RR396 pKa = 11.84GQII399 pKa = 3.58
Molecular weight: 47.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1389 |
77 |
548 |
277.8 |
31.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.479 ± 1.306 | 1.368 ± 0.404 |
4.896 ± 1.011 | 7.631 ± 1.562 |
4.536 ± 0.376 | 6.551 ± 1.251 |
1.512 ± 0.246 | 6.695 ± 0.49 |
9.719 ± 2.68 | 7.415 ± 0.72 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.52 ± 0.391 | 6.911 ± 0.77 |
3.6 ± 0.925 | 3.6 ± 0.523 |
3.744 ± 0.533 | 5.688 ± 1.15 |
7.919 ± 0.928 | 3.384 ± 0.976 |
1.152 ± 0.423 | 4.68 ± 0.424 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
