
Cryptococcus gattii serotype B (strain R265) (Filobasidiella gattii) (Cryptococcus bacillisporus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Cryptococcaceae; Cryptococcus; Cryptococcus gattii species complex; Cryptococcus gattii VGII
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
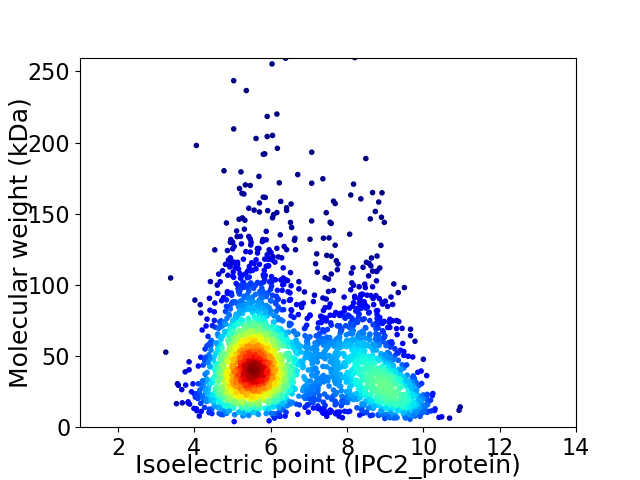
Virtual 2D-PAGE plot for 2942 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A095CAX8|A0A095CAX8_CRYGR Selenoprotein O OS=Cryptococcus gattii serotype B (strain R265) OX=294750 GN=CNBG_2597 PE=3 SV=1
MM1 pKa = 7.68IPIRR5 pKa = 11.84QTIPFGLLSCLALLGLVKK23 pKa = 10.66ADD25 pKa = 3.27GSTSNTSWPEE35 pKa = 3.58WVTNDD40 pKa = 3.3YY41 pKa = 11.4DD42 pKa = 4.31CVIGCMSGFNDD53 pKa = 4.73TITYY57 pKa = 9.86IPQDD61 pKa = 3.6ALAQQAFDD69 pKa = 4.54CTSSTCKK76 pKa = 10.64GDD78 pKa = 3.29VTGNYY83 pKa = 9.07YY84 pKa = 8.29QTLYY88 pKa = 10.8YY89 pKa = 9.78IQLFYY94 pKa = 10.83ATGSIYY100 pKa = 10.17EE101 pKa = 4.32YY102 pKa = 10.86SDD104 pKa = 3.6SAPDD108 pKa = 4.26GYY110 pKa = 11.29KK111 pKa = 10.14HH112 pKa = 7.21ANFTDD117 pKa = 3.96STGTTSDD124 pKa = 3.55AAVSGQSGDD133 pKa = 4.64DD134 pKa = 3.59NTIDD138 pKa = 3.51PTDD141 pKa = 3.3TSEE144 pKa = 5.39ADD146 pKa = 3.32FASATDD152 pKa = 4.12GVKK155 pKa = 7.93TTGSADD161 pKa = 3.29GGNDD165 pKa = 3.41GSSTDD170 pKa = 3.56TAAVAVDD177 pKa = 3.53TGRR180 pKa = 11.84EE181 pKa = 3.97NNGAAVKK188 pKa = 10.46ASEE191 pKa = 4.56PDD193 pKa = 3.78SIISGSEE200 pKa = 3.73PFSTKK205 pKa = 9.17STSSHH210 pKa = 6.17TDD212 pKa = 2.83TSIASSSVAADD223 pKa = 3.33ISDD226 pKa = 3.4SGAFKK231 pKa = 10.61IIGGGVWQSVLGALGALVLGGVWVGMM257 pKa = 4.33
MM1 pKa = 7.68IPIRR5 pKa = 11.84QTIPFGLLSCLALLGLVKK23 pKa = 10.66ADD25 pKa = 3.27GSTSNTSWPEE35 pKa = 3.58WVTNDD40 pKa = 3.3YY41 pKa = 11.4DD42 pKa = 4.31CVIGCMSGFNDD53 pKa = 4.73TITYY57 pKa = 9.86IPQDD61 pKa = 3.6ALAQQAFDD69 pKa = 4.54CTSSTCKK76 pKa = 10.64GDD78 pKa = 3.29VTGNYY83 pKa = 9.07YY84 pKa = 8.29QTLYY88 pKa = 10.8YY89 pKa = 9.78IQLFYY94 pKa = 10.83ATGSIYY100 pKa = 10.17EE101 pKa = 4.32YY102 pKa = 10.86SDD104 pKa = 3.6SAPDD108 pKa = 4.26GYY110 pKa = 11.29KK111 pKa = 10.14HH112 pKa = 7.21ANFTDD117 pKa = 3.96STGTTSDD124 pKa = 3.55AAVSGQSGDD133 pKa = 4.64DD134 pKa = 3.59NTIDD138 pKa = 3.51PTDD141 pKa = 3.3TSEE144 pKa = 5.39ADD146 pKa = 3.32FASATDD152 pKa = 4.12GVKK155 pKa = 7.93TTGSADD161 pKa = 3.29GGNDD165 pKa = 3.41GSSTDD170 pKa = 3.56TAAVAVDD177 pKa = 3.53TGRR180 pKa = 11.84EE181 pKa = 3.97NNGAAVKK188 pKa = 10.46ASEE191 pKa = 4.56PDD193 pKa = 3.78SIISGSEE200 pKa = 3.73PFSTKK205 pKa = 9.17STSSHH210 pKa = 6.17TDD212 pKa = 2.83TSIASSSVAADD223 pKa = 3.33ISDD226 pKa = 3.4SGAFKK231 pKa = 10.61IIGGGVWQSVLGALGALVLGGVWVGMM257 pKa = 4.33
Molecular weight: 26.4 kDa
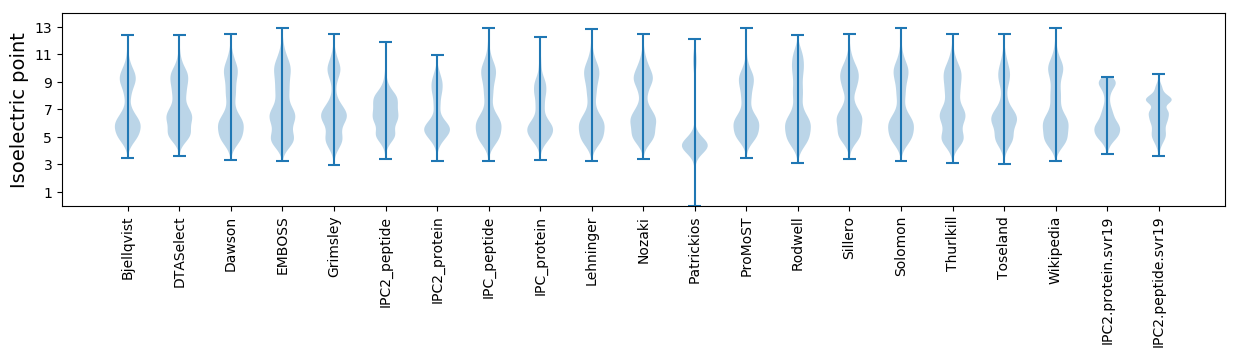
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A095C319|A0A095C319_CRYGR Calcineurin-binding protein OS=Cryptococcus gattii serotype B (strain R265) OX=294750 GN=CNBG_1133 PE=3 SV=1
MM1 pKa = 7.43SPASPKK7 pKa = 9.93QPHH10 pKa = 6.4SPTTPPSPTSSPTHH24 pKa = 5.89SILRR28 pKa = 11.84RR29 pKa = 11.84TASTLSAPSHH39 pKa = 5.19SHH41 pKa = 5.76KK42 pKa = 10.96KK43 pKa = 8.34MLRR46 pKa = 11.84FTQVHH51 pKa = 6.56RR52 pKa = 11.84DD53 pKa = 3.23SDD55 pKa = 4.24TLLSTVGHH63 pKa = 6.29NEE65 pKa = 3.45DD66 pKa = 3.58VYY68 pKa = 9.68YY69 pKa = 10.23QGRR72 pKa = 11.84TVEE75 pKa = 4.18EE76 pKa = 3.93GLYY79 pKa = 9.78TDD81 pKa = 5.23RR82 pKa = 11.84LATDD86 pKa = 3.6RR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84YY90 pKa = 8.56HH91 pKa = 6.31QSDD94 pKa = 3.46PGTPNFQEE102 pKa = 3.92TADD105 pKa = 3.7QIRR108 pKa = 11.84RR109 pKa = 11.84TLSLASVDD117 pKa = 4.1SLLLLSVNQSEE128 pKa = 4.47RR129 pKa = 11.84TKK131 pKa = 10.93FLAEE135 pKa = 3.7VSRR138 pKa = 11.84AGRR141 pKa = 11.84GLVWRR146 pKa = 11.84DD147 pKa = 3.18EE148 pKa = 4.45SEE150 pKa = 4.23QCLLPQDD157 pKa = 3.74NEE159 pKa = 4.33RR160 pKa = 11.84AVVLALKK167 pKa = 10.4RR168 pKa = 11.84GLRR171 pKa = 11.84SFILAFSVRR180 pKa = 11.84SGINVLLLLFRR191 pKa = 11.84LLRR194 pKa = 11.84KK195 pKa = 9.05HH196 pKa = 6.22APKK199 pKa = 10.65LSLPLLRR206 pKa = 11.84HH207 pKa = 6.6AIFGPEE213 pKa = 3.92PFRR216 pKa = 11.84FGGMIGTFTLLYY228 pKa = 10.32TFTLHH233 pKa = 7.65LLRR236 pKa = 11.84LAPPLSYY243 pKa = 10.16FRR245 pKa = 11.84RR246 pKa = 11.84RR247 pKa = 11.84VRR249 pKa = 11.84AGLWNKK255 pKa = 7.25TTFGPPEE262 pKa = 3.88RR263 pKa = 11.84EE264 pKa = 4.07GNEE267 pKa = 4.16GEE269 pKa = 4.55RR270 pKa = 11.84RR271 pKa = 11.84WQAAVAGAVGSLGLLWEE288 pKa = 4.88GKK290 pKa = 7.45SRR292 pKa = 11.84RR293 pKa = 11.84RR294 pKa = 11.84GVAQQMFVRR303 pKa = 11.84GLQASYY309 pKa = 10.87NQYY312 pKa = 8.55TPRR315 pKa = 11.84FGIHH319 pKa = 6.45IPHH322 pKa = 7.38GDD324 pKa = 3.57LLLFGACCGQIMFAFLMSPEE344 pKa = 4.79TIPKK348 pKa = 9.8EE349 pKa = 3.94YY350 pKa = 10.98NNWILAASRR359 pKa = 11.84VPSFAILANRR369 pKa = 11.84TQVRR373 pKa = 11.84QGYY376 pKa = 8.33IQDD379 pKa = 4.02ALVQEE384 pKa = 4.14ALNYY388 pKa = 10.36KK389 pKa = 10.28GITAINRR396 pKa = 11.84RR397 pKa = 11.84RR398 pKa = 11.84LEE400 pKa = 3.86NVLQKK405 pKa = 10.25IRR407 pKa = 11.84GGQQPDD413 pKa = 3.45AVPCGMVHH421 pKa = 6.64PWCNSCTEE429 pKa = 4.14TNFRR433 pKa = 11.84RR434 pKa = 11.84FFAVCRR440 pKa = 11.84FMLPVYY446 pKa = 10.18SALHH450 pKa = 6.75LIPMLVLRR458 pKa = 11.84RR459 pKa = 11.84HH460 pKa = 5.03HH461 pKa = 6.36VKK463 pKa = 10.27RR464 pKa = 11.84DD465 pKa = 3.6PVRR468 pKa = 11.84MMARR472 pKa = 11.84ALWGIGRR479 pKa = 11.84SCSFLGVFVVIYY491 pKa = 10.02QVLFCARR498 pKa = 11.84VQALEE503 pKa = 4.07KK504 pKa = 10.94SRR506 pKa = 11.84GSDD509 pKa = 4.35FIRR512 pKa = 11.84KK513 pKa = 8.46LLKK516 pKa = 10.17RR517 pKa = 11.84KK518 pKa = 8.95EE519 pKa = 4.22VFWALGFTTCLSLLVEE535 pKa = 4.29EE536 pKa = 4.78KK537 pKa = 10.45KK538 pKa = 10.69RR539 pKa = 11.84RR540 pKa = 11.84AEE542 pKa = 3.54LAMYY546 pKa = 8.07VLPRR550 pKa = 11.84ALEE553 pKa = 4.33SAWSGARR560 pKa = 11.84KK561 pKa = 9.28RR562 pKa = 11.84AWVPLVPFGEE572 pKa = 4.74TILGAAAMAMVMDD585 pKa = 5.41AYY587 pKa = 9.89KK588 pKa = 10.08HH589 pKa = 5.81QPDD592 pKa = 3.71AMSGIVRR599 pKa = 11.84RR600 pKa = 11.84LLFQLVGPGG609 pKa = 3.09
MM1 pKa = 7.43SPASPKK7 pKa = 9.93QPHH10 pKa = 6.4SPTTPPSPTSSPTHH24 pKa = 5.89SILRR28 pKa = 11.84RR29 pKa = 11.84TASTLSAPSHH39 pKa = 5.19SHH41 pKa = 5.76KK42 pKa = 10.96KK43 pKa = 8.34MLRR46 pKa = 11.84FTQVHH51 pKa = 6.56RR52 pKa = 11.84DD53 pKa = 3.23SDD55 pKa = 4.24TLLSTVGHH63 pKa = 6.29NEE65 pKa = 3.45DD66 pKa = 3.58VYY68 pKa = 9.68YY69 pKa = 10.23QGRR72 pKa = 11.84TVEE75 pKa = 4.18EE76 pKa = 3.93GLYY79 pKa = 9.78TDD81 pKa = 5.23RR82 pKa = 11.84LATDD86 pKa = 3.6RR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84YY90 pKa = 8.56HH91 pKa = 6.31QSDD94 pKa = 3.46PGTPNFQEE102 pKa = 3.92TADD105 pKa = 3.7QIRR108 pKa = 11.84RR109 pKa = 11.84TLSLASVDD117 pKa = 4.1SLLLLSVNQSEE128 pKa = 4.47RR129 pKa = 11.84TKK131 pKa = 10.93FLAEE135 pKa = 3.7VSRR138 pKa = 11.84AGRR141 pKa = 11.84GLVWRR146 pKa = 11.84DD147 pKa = 3.18EE148 pKa = 4.45SEE150 pKa = 4.23QCLLPQDD157 pKa = 3.74NEE159 pKa = 4.33RR160 pKa = 11.84AVVLALKK167 pKa = 10.4RR168 pKa = 11.84GLRR171 pKa = 11.84SFILAFSVRR180 pKa = 11.84SGINVLLLLFRR191 pKa = 11.84LLRR194 pKa = 11.84KK195 pKa = 9.05HH196 pKa = 6.22APKK199 pKa = 10.65LSLPLLRR206 pKa = 11.84HH207 pKa = 6.6AIFGPEE213 pKa = 3.92PFRR216 pKa = 11.84FGGMIGTFTLLYY228 pKa = 10.32TFTLHH233 pKa = 7.65LLRR236 pKa = 11.84LAPPLSYY243 pKa = 10.16FRR245 pKa = 11.84RR246 pKa = 11.84RR247 pKa = 11.84VRR249 pKa = 11.84AGLWNKK255 pKa = 7.25TTFGPPEE262 pKa = 3.88RR263 pKa = 11.84EE264 pKa = 4.07GNEE267 pKa = 4.16GEE269 pKa = 4.55RR270 pKa = 11.84RR271 pKa = 11.84WQAAVAGAVGSLGLLWEE288 pKa = 4.88GKK290 pKa = 7.45SRR292 pKa = 11.84RR293 pKa = 11.84RR294 pKa = 11.84GVAQQMFVRR303 pKa = 11.84GLQASYY309 pKa = 10.87NQYY312 pKa = 8.55TPRR315 pKa = 11.84FGIHH319 pKa = 6.45IPHH322 pKa = 7.38GDD324 pKa = 3.57LLLFGACCGQIMFAFLMSPEE344 pKa = 4.79TIPKK348 pKa = 9.8EE349 pKa = 3.94YY350 pKa = 10.98NNWILAASRR359 pKa = 11.84VPSFAILANRR369 pKa = 11.84TQVRR373 pKa = 11.84QGYY376 pKa = 8.33IQDD379 pKa = 4.02ALVQEE384 pKa = 4.14ALNYY388 pKa = 10.36KK389 pKa = 10.28GITAINRR396 pKa = 11.84RR397 pKa = 11.84RR398 pKa = 11.84LEE400 pKa = 3.86NVLQKK405 pKa = 10.25IRR407 pKa = 11.84GGQQPDD413 pKa = 3.45AVPCGMVHH421 pKa = 6.64PWCNSCTEE429 pKa = 4.14TNFRR433 pKa = 11.84RR434 pKa = 11.84FFAVCRR440 pKa = 11.84FMLPVYY446 pKa = 10.18SALHH450 pKa = 6.75LIPMLVLRR458 pKa = 11.84RR459 pKa = 11.84HH460 pKa = 5.03HH461 pKa = 6.36VKK463 pKa = 10.27RR464 pKa = 11.84DD465 pKa = 3.6PVRR468 pKa = 11.84MMARR472 pKa = 11.84ALWGIGRR479 pKa = 11.84SCSFLGVFVVIYY491 pKa = 10.02QVLFCARR498 pKa = 11.84VQALEE503 pKa = 4.07KK504 pKa = 10.94SRR506 pKa = 11.84GSDD509 pKa = 4.35FIRR512 pKa = 11.84KK513 pKa = 8.46LLKK516 pKa = 10.17RR517 pKa = 11.84KK518 pKa = 8.95EE519 pKa = 4.22VFWALGFTTCLSLLVEE535 pKa = 4.29EE536 pKa = 4.78KK537 pKa = 10.45KK538 pKa = 10.69RR539 pKa = 11.84RR540 pKa = 11.84AEE542 pKa = 3.54LAMYY546 pKa = 8.07VLPRR550 pKa = 11.84ALEE553 pKa = 4.33SAWSGARR560 pKa = 11.84KK561 pKa = 9.28RR562 pKa = 11.84AWVPLVPFGEE572 pKa = 4.74TILGAAAMAMVMDD585 pKa = 5.41AYY587 pKa = 9.89KK588 pKa = 10.08HH589 pKa = 5.81QPDD592 pKa = 3.71AMSGIVRR599 pKa = 11.84RR600 pKa = 11.84LLFQLVGPGG609 pKa = 3.09
Molecular weight: 68.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1331672 |
38 |
2365 |
452.6 |
49.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.515 ± 0.042 | 1.0 ± 0.016 |
5.322 ± 0.028 | 6.754 ± 0.043 |
3.43 ± 0.029 | 7.186 ± 0.038 |
2.302 ± 0.019 | 4.888 ± 0.032 |
5.422 ± 0.04 | 8.964 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.148 ± 0.017 | 3.377 ± 0.021 |
6.458 ± 0.053 | 3.766 ± 0.036 |
5.848 ± 0.034 | 8.795 ± 0.064 |
5.745 ± 0.026 | 6.157 ± 0.03 |
1.362 ± 0.016 | 2.56 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
